Mutation history of the roma/gypsies - PubMed (original) (raw)
. 2004 Oct;75(4):596-609.
doi: 10.1086/424759. Epub 2004 Aug 20.
David Gresham, Dora Angelicheva, Ivailo Tournev, Rebecca Gooding, Velina Guergueltcheva, Carolin Schmidt, Angela Abicht, Hanns Lochmuller, Attila Tordai, Lajos Kalmar, Melinda Nagy, Veronika Karcagi, Marc Jeanpierre, Agnes Herczegfalvi, David Beeson, Viswanathan Venkataraman, Kim Warwick Carter, Jeff Reeve, Rosario de Pablo, Vaidutis Kucinskas, Luba Kalaydjieva
Affiliations
- PMID: 15322984
- PMCID: PMC1182047
- DOI: 10.1086/424759
Mutation history of the roma/gypsies
Bharti Morar et al. Am J Hum Genet. 2004 Oct.
Abstract
The 8-10 million European Roma/Gypsies are a founder population of common origins that has subsequently split into multiple socially divergent and geographically dispersed Gypsy groups. Unlike other founder populations, whose genealogy has been extensively documented, the demographic history of the Gypsies is not fully understood and, given the lack of written records, has to be inferred from current genetic data. In this study, we have used five disease loci harboring private Gypsy mutations to examine some missing historical parameters and current structure. We analyzed the frequency distribution of the five mutations in 832-1,363 unrelated controls, representing 14 Gypsy populations, and the diversification of chromosomal haplotypes in 501 members of affected families. Sharing of mutations and high carrier rates supported a strong founder effect, and the identity of the congenital myasthenia 1267delG mutation in Gypsy and Indian/Pakistani chromosomes provided the best evidence yet of the Indian origins of the Gypsies. However, dramatic differences in mutation frequencies and haplotype divergence and very limited haplotype sharing pointed to strong internal differentiation and characterized the Gypsies as a founder population comprising multiple subisolates. Using disease haplotype coalescence times at the different loci, we estimated that the entire Gypsy population was founded approximately 32-40 generations ago, with secondary and tertiary founder events occurring approximately 16-25 generations ago. The existence of multiple subisolates, with endogamy maintained to the present day, suggests a general approach to complex disorders in which initial gene mapping could be performed in large families from a single Gypsy group, whereas fine mapping would rely on the informed sampling of the divergent subisolates and searching for the shared genomic region that displays the strongest linkage disequilibrium with the disease.
Figures
Figure 1
Assignment of participating subjects to migrational/linguistic categories and individual Gypsy groups within the categories. The Xoroxane and Dassikane (indicated with an asterisk [*]) do not specify individual Gypsy groups but represent broader cultural anthropological divisions within the Balkan category, in which group identity has been lost. The western European migrational category comprises subjects from Hungary, Slovenia, the Czech Republic, Lithuania, Germany, France, Italy, Spain, and Portugal.
Figure 2
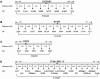
Genetic maps used in the haplotype analysis of four of the founder mutations. Markers are shown as equidistant from one another. The position of the mutation is indicated with a black square. Markers designated with an “S” number are known microsatellites found in public databases. Microsatellites identified during positional cloning studies include SLAP (NDRG1 region), 23090ta1, 1908ca1, 21594at1, 68530gt1, 68530ca1, and 68530gt2 (CTDP1 region). LOC125267, LOC125261, DIM1, and PAR6 in this region represent insertion/deletion polymorphisms. Markers in boldface type were included in the comparative analysis of regions spanning ∼3 cM around each disease mutation. A, CHRNE on 17p13.2. B, NDRG1 on 8q24.22. C, SGCG on 13q12.12. _D, CTDP1_on 18qter.
Figure 3
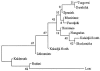
Neighbor-joining tree of Gypsy groups, based on the gene frequency data obtained from the population screening of the five disease mutations and Nei’s standard genetic distance (Nei 1987). The tree was generated using the DISPAN package (Ota 1993), on the basis of the neighbor-joining method (Saitou and Nei 1987). The numbers indicate the robustness of the branches in the tree, assessed with a bootstrap approach.
Figure 4
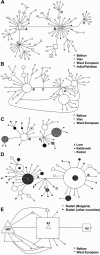
Networks showing the genealogical relationships of disease haplotypes. The size of the symbols is proportional to the frequency of each haplotype. A, CMS haplotypes in the CHRNE region. B, HMSNL haplotypes in the NDRG1 region. C, HMSNL haplotypes in Gypsy groups from the Vlax migrational category. D, CCFDN haplotypes in the CTDP1 region. E, LGMD2C haplotypes in the SGCG region. For CMS and HMSNL, only haplotype groups comprising more than five unique haplotypes are shown.
Figure 5
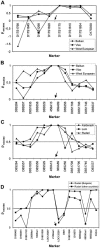
LD estimated by pexcess (Hastbacka et al. 1992, 1994) for markers surrounding the founder disease mutations. Markers are shown as equidistant from one another. Intermarker distances are shown in figures 2 and 6. The arrows indicate the approximate location of the disease mutation. A, LD in the CMS gene region in the three migrational categories. B, LD in the HMSNL region in the three migrational categories. C, LD in the HMSNL region in Gypsy groups from the Vlax migrational category. D, LD in the CCFDN region for geographically separated Rudari groups.
Figure 6
Pairwise LD between markers in the HMSNL and CCFDN disease gene regions in four Gypsy groups. A, Rudari. B, Kalderash. C, Lom. D, Turgovzi. Marker loci are listed across the bottom and down the left side. A scale indicating physical distances (in kb) between markers is shown at the bottom. _D_′ values are shown in the upper left-hand triangle, and P values are shown in the lower right-hand triangle. The shading patterns used to illustrate the scale of _D_′ and P values are shown above and at the right-hand side of the top panel. P values <.05 have been corrected for multiple comparisons through use of the Holm-Sidak approach (Lautenberger et al. 2000).
Similar articles
- Carrier rates of the ancestral Indian W24X mutation in GJB2 in the general Gypsy population and individual subisolates.
Bouwer S, Angelicheva D, Chandler D, Seeman P, Tournev I, Kalaydjieva L. Bouwer S, et al. Genet Test. 2007 Winter;11(4):455-8. doi: 10.1089/gte.2007.0048. Genet Test. 2007. PMID: 18294064 - A newly discovered founder population: the Roma/Gypsies.
Kalaydjieva L, Morar B, Chaix R, Tang H. Kalaydjieva L, et al. Bioessays. 2005 Oct;27(10):1084-94. doi: 10.1002/bies.20287. Bioessays. 2005. PMID: 16163730 - Origins and divergence of the Roma (gypsies).
Gresham D, Morar B, Underhill PA, Passarino G, Lin AA, Wise C, Angelicheva D, Calafell F, Oefner PJ, Shen P, Tournev I, de Pablo R, Kuĉinskas V, Perez-Lezaun A, Marushiakova E, Popov V, Kalaydjieva L. Gresham D, et al. Am J Hum Genet. 2001 Dec;69(6):1314-31. doi: 10.1086/324681. Epub 2001 Nov 9. Am J Hum Genet. 2001. PMID: 11704928 Free PMC article. - Neuromuscular disorders in the Gypsy ethnic group. A short review.
Navarro C, Teijeira S. Navarro C, et al. Acta Myol. 2003 May;22(1):11-4. Acta Myol. 2003. PMID: 12966699 Review. - Population Genetics of the European Roma-A Review.
Ena GF, Aizpurua-Iraola J, Font-Porterias N, Calafell F, Comas D. Ena GF, et al. Genes (Basel). 2022 Nov 8;13(11):2068. doi: 10.3390/genes13112068. Genes (Basel). 2022. PMID: 36360305 Free PMC article. Review.
Cited by
- Phenotypic Variability of LGMD 2C/R5 in a Genetically Homogenous Group of Bulgarian Muslim Roma.
Taneva A, Gresham D, Guergueltcheva V, Chamova T, Bojinova V, Gospodinova M, Katzarova M, Petkov R, Voit T, Aneva L, Asenov O, Georgieva B, Mihaylova V, Bichev S, Todorov T, Todorova A, Kalaydjieva L, Tournev I. Taneva A, et al. Genes (Basel). 2024 Aug 30;15(9):1144. doi: 10.3390/genes15091144. Genes (Basel). 2024. PMID: 39336735 Free PMC article. - Characterization of Clinical Phenotypes in Congenital Myasthenic Syndrome Associated with the c.1327delG Frameshift Mutation in CHRNE Encoding the Acetylcholine Receptor Epsilon Subunit.
Kastreva K, Chamova T, Blagoeva S, Bichev S, Mihaylova V, Meyer S, Thompson R, Cherninkova S, Guergueltcheva V, Lochmuller H, Tournev I. Kastreva K, et al. J Neuromuscul Dis. 2024;11(5):1011-1020. doi: 10.3233/JND-230235. J Neuromuscul Dis. 2024. PMID: 38995797 Free PMC article. - Founder mutations and rare disease in the Arab world.
Marafi D. Marafi D. Dis Model Mech. 2024 Jun 1;17(6):dmm050715. doi: 10.1242/dmm.050715. Epub 2024 Jun 26. Dis Model Mech. 2024. PMID: 38922202 Free PMC article. Review. - Population history modulates the fitness effects of Copy Number Variation in the Roma.
Antinucci M, Comas D, Calafell F. Antinucci M, et al. Hum Genet. 2023 Sep;142(9):1327-1343. doi: 10.1007/s00439-023-02579-5. Epub 2023 Jun 14. Hum Genet. 2023. PMID: 37311904 Free PMC article. - Pathogenic Variants of SLC22A12 (URAT1) and SLC2A9 (GLUT9) in Spanish Patients with Renal Hypouricemia: Founder Effect of SLC2A9 Variant c.374C>T; p.(T125M).
Perdomo-Ramirez A, Cordoba-Lanus E, Trujillo-Frias CJ, Gonzalez-Navasa C, Ramos-Trujillo E, Luis-Yanes MI, Garcia-Nieto V, Claverie-Martin F; RenalTube. Perdomo-Ramirez A, et al. Int J Mol Sci. 2023 May 8;24(9):8455. doi: 10.3390/ijms24098455. Int J Mol Sci. 2023. PMID: 37176161 Free PMC article.
References
Electronic-Database Information
- National Center for Biotechnology Information Genome View, http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=9606 (for the deCODE genetic map)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HMSNL, CCFDN, CMS, LGMD2C, and galactokinase deficiency)
References
- Abicht A, Stucka R, Karcagi V, Herczegfalvi A, Horvath R, Mortier W, Schara U, Ramaekers V, Jost W, Brunner J, Janssen G, Seidel U, Schlotter B, Muller-Felber W, Pongratz D, Rudel R, Lochmuller H (1999) A common mutation (epsilon1267delG) in congenital myasthenic patients of Gypsy ethnic origin. Neurology 53:1564–1569 - PubMed
- Angelicheva D, Turnev I, Dye D, Chandler D, Thomas PK, Kalaydjieva L (1999) Congenital cataracts facial dysmorphism neuropathy (CCFDN) syndrome: a novel developmental disorder in Gypsies maps to 18qter. Eur J Hum Genet 7:560–566 - PubMed
- Angius A, Bebbere D, Petretto E, Falchi M, Forabosco P, Maestrale B, Casu G, Persico I, Melis PM, Pirastu M (2002) Not all isolates are equal: linkage disequilibrium analysis on Xq13.3 reveals different patterns in Sardinian sub-populations. Hum Genet 111:9–1510.1007/s00439-002-0753-z - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources