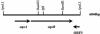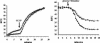Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli - PubMed (original) (raw)
Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli
Lars Hestbjerg Hansen et al. Antimicrob Agents Chemother. 2004 Sep.
Abstract
We report here the first gene-encoded resistance mechanism to the swine growth enhancer olaquindox. The genetic elements involved in resistance to olaquindox were subcloned and sequenced from a conjugative plasmid isolated from Escherichia coli. The subcloned fragment contained two open reading frames, oqxA and oqxB, that are homologous to several resistance-nodulation-cell-division family efflux systems from different species. The putative protein sequences were aligned to both experimentally verified and putative efflux pumps. We show that oqxA and oqxB are expressed in E. coli. Plasmids containing the oqxAB genes yielded high (>128 microg/ml) resistance to olaquindox in E. coli, whereas strains containing the control plasmid showed low resistance to the drug (8 microg/ml). The oqxAB-encoded pump also conferred high (>64 microg/ml) resistance to chloramphenicol. We demonstrate that the subcloned fragment conferred H(+)-dependent ethidium efflux abilities to E. coli strain N43. In addition, we show that the efflux system is dependent on the host TolC outer membrane protein when expressed in E. coli.
Figures
FIG. 1.
The 6-kb ApaLI insert from pOLA52 revealed three ORFs, and their locations are indicated in the diagram. The oqxA and oqxB genes are situated on the plus strand, ORF3 is located on the minus strand. Some selected restriction sites are indicated.
FIG. 2.
(A) Comparison of the putative protein sequence of OqxA and the entire sequences of the E. coli AcrA, X. axonopodis MexE, and P. aeruginosa MexX proteins. (B) Comparison of the putative protein sequence of OqxB and the entire sequences of the E. coli AcrB, X. axonopodis MexF, and P. aeruginosa MexY proteins. Black letters on a white background indicate different amino acid residues. Shaded black letters indicate similar residues. White letters on a black background indicate identical residues.
FIG. 2.
(A) Comparison of the putative protein sequence of OqxA and the entire sequences of the E. coli AcrA, X. axonopodis MexE, and P. aeruginosa MexX proteins. (B) Comparison of the putative protein sequence of OqxB and the entire sequences of the E. coli AcrB, X. axonopodis MexF, and P. aeruginosa MexY proteins. Black letters on a white background indicate different amino acid residues. Shaded black letters indicate similar residues. White letters on a black background indicate identical residues.
FIG. 2.
(A) Comparison of the putative protein sequence of OqxA and the entire sequences of the E. coli AcrA, X. axonopodis MexE, and P. aeruginosa MexX proteins. (B) Comparison of the putative protein sequence of OqxB and the entire sequences of the E. coli AcrB, X. axonopodis MexF, and P. aeruginosa MexY proteins. Black letters on a white background indicate different amino acid residues. Shaded black letters indicate similar residues. White letters on a black background indicate identical residues.
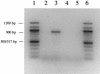
FIG. 3.
RT-PCR analysis of oqxAB operon and agarose gel electrophoresis of RT-PCR products. Products were visualized in a 1.2% agarose gel. Lanes 1 and 6, 100-bp marker; lanes 2 and 3, RT-PCRs with N43 (lane 2) or N43/pOLA52 (lane 3) as a template; lanes 4 and 5, control PCRs on templates of total RNA from N43 (lane 4) or N43/pOLA52 (lane 5).
FIG. 4.
(Left panel) Uptake of ethidium in E. coli N43 cells containing either pLOW2 (○) or pLOW2::oqxAB (▵). Cells were exposed to ethidium bromide at 0 min. CCCP was added to the cells after 9 min (point indicated by arrow). The fluorescence of cells (shown as relative fluorescence units [RFU]), caused by the presence of ethidium, was measured continuously during the assay. (Right panel) Ethidium efflux. Starved cells of N43 cells containing either pLOW2 (○) or pLOW2::oqxAB (▵) were loaded with ethidium bromide for 1 h prior to the start of the assay. At 5 min after assay start, glucose and thiamine were added to energize the cells (indicated by arrow). The fluorescence of cells (shown as relative fluorescence units [RFU]), caused by the presence of ethidium, was measured continuously during the assay.
Similar articles
- The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs.
Hansen LH, Sørensen SJ, Jørgensen HS, Jensen LB. Hansen LH, et al. Microb Drug Resist. 2005 Winter;11(4):378-82. doi: 10.1089/mdr.2005.11.378. Microb Drug Resist. 2005. PMID: 16359198 - Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria.
Hansen LH, Jensen LB, Sørensen HI, Sørensen SJ. Hansen LH, et al. J Antimicrob Chemother. 2007 Jul;60(1):145-7. doi: 10.1093/jac/dkm167. Epub 2007 May 24. J Antimicrob Chemother. 2007. PMID: 17526501 - Efflux pump genes of the resistance-nodulation-division family in Burkholderia cenocepacia genome.
Guglierame P, Pasca MR, De Rossi E, Buroni S, Arrigo P, Manina G, Riccardi G. Guglierame P, et al. BMC Microbiol. 2006 Jul 20;6:66. doi: 10.1186/1471-2180-6-66. BMC Microbiol. 2006. PMID: 16857052 Free PMC article. - [The role of cell wall organization and active efflux pump systems in multidrug resistance of bacteria].
Hasdemir U. Hasdemir U. Mikrobiyol Bul. 2007 Apr;41(2):309-27. Mikrobiyol Bul. 2007. PMID: 17682720 Review. Turkish. - The multiple antibiotic resistance (mar) locus and its significance.
Randall LP, Woodward MJ. Randall LP, et al. Res Vet Sci. 2002 Apr;72(2):87-93. doi: 10.1053/rvsc.2001.0537. Res Vet Sci. 2002. PMID: 12027588 Review.
Cited by
- Genomic Analysis of the Emergence and Rapid Global Dissemination of the Clonal Group 258 Klebsiella pneumoniae Pandemic.
Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, de Man T, Rasheed JK, Engelthaler DM, Keim P, Limbago BM. Bowers JR, et al. PLoS One. 2015 Jul 21;10(7):e0133727. doi: 10.1371/journal.pone.0133727. eCollection 2015. PLoS One. 2015. PMID: 26196384 Free PMC article. - Development of quinoxaline 1, 4-dioxides resistance in Escherichia coli and molecular change under resistance selection.
Guo W, Hao H, Dai M, Wang Y, Huang L, Peng D, Wang X, Wang H, Yao M, Sun Y, Liu Z, Yuan Z. Guo W, et al. PLoS One. 2012;7(8):e43322. doi: 10.1371/journal.pone.0043322. Epub 2012 Aug 28. PLoS One. 2012. PMID: 22952665 Free PMC article. - Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies.
Lupo A, Coyne S, Berendonk TU. Lupo A, et al. Front Microbiol. 2012 Jan 26;3:18. doi: 10.3389/fmicb.2012.00018. eCollection 2012. Front Microbiol. 2012. PMID: 22303296 Free PMC article. - New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate.
Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H, Shibayama K, Konda T, Arakawa Y. Yamane K, et al. Antimicrob Agents Chemother. 2007 Sep;51(9):3354-60. doi: 10.1128/AAC.00339-07. Epub 2007 Jun 4. Antimicrob Agents Chemother. 2007. PMID: 17548499 Free PMC article. - Sequencing of IncX-plasmids suggests ubiquity of mobile forms of a biofilm-promoting gene cassette recruited from Klebsiella pneumoniae.
Burmølle M, Norman A, Sørensen SJ, Hansen LH. Burmølle M, et al. PLoS One. 2012;7(7):e41259. doi: 10.1371/journal.pone.0041259. Epub 2012 Jul 23. PLoS One. 2012. PMID: 22844447 Free PMC article.
References
- DANMAP. 1999. Resistance monitoring in Denmark, 1997-DANMAP. Wkly Epidemiol. Rec. 74:125-127. - PubMed
- da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases