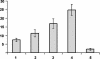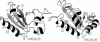A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana - PubMed (original) (raw)
. 2004 Sep 7;101(36):13380-5.
doi: 10.1073/pnas.0405248101. Epub 2004 Aug 25.
Marco da Costa, Lieven De Veylder, Frédérique Dewitte, Klaas Vandepoele, Sahar Hassan, Jean-Michel Wieruszeski, Florence Corellou, Jean-Denis Faure, Marc Van Montagu, Dirk Inzé, Guy Lippens
Affiliations
- PMID: 15329414
- PMCID: PMC516575
- DOI: 10.1073/pnas.0405248101
A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana
Isabelle Landrieu et al. Proc Natl Acad Sci U S A. 2004.
Erratum in
- Proc Natl Acad Sci U S A. 2004 Nov 16;101(46):16391
Abstract
The dual-specificity CDC25 phosphatases are critical positive regulators of cyclin-dependent kinases (CDKs). Even though an antagonistic Arabidopsis thaliana WEE1 kinase has been cloned and tyrosine phosphorylation of its CDKs has been demonstrated, no valid candidate for a CDC25 protein has been reported in higher plants. We identify a CDC25-related protein (Arath;CDC25) of A. thaliana, constituted by a sole catalytic domain. The protein has a tyrosine-phosphatase activity and stimulates the kinase activity of Arabidopsis CDKs. Its tertiary structure was obtained by NMR spectroscopy and confirms that Arath;CDC25 belongs structurally to the classical CDC25 superfamily with a central five-stranded beta-sheet surrounded by helices. A particular feature of the protein, however, is the presence of an additional zinc-binding loop in the C-terminal part. NMR mapping studies revealed the interaction with phosphorylated peptidic models derived from the conserved CDK loop containing the phosphothreonine-14 and phosphotyrosine-15. We conclude that despite sequence divergence, Arath;CDC25 is structurally and functionally an isoform of the CDC25 superfamily, which is conserved in yeast and in plants, including Arabidopsis and rice.
Figures
Fig. 1.
Sequence comparison of the Arath;CDC25 protein with CDC25, small CDC25-like tyrosine phosphatases, and the yeast arsenate reductase ACR2. Identical amino acids in all of the sequences are shown in white on a black background. Conserved amino acids are in bold. Secondary structure elements are indicated above the alignment for human CDC25A (PDB ID 1C25) and below for A. thaliana Arath;CDC25. The catalytic loop is indicated by the HC(X)5R motif. CDC25ACDC25A_human, human CDC25A (accession number NP_001780); CDC25B_human, human CDC25B (NP_068658); CDC25C_human, human CDC25C (NP_073720); CDC25_pombe, Sch. pombe CDC25 (NP_013750); MIH1_cerevisiae, S. cerevisiae CDC25 (NP_013750); YGR203_cerevisiae, S. cerevisiae protein of unknown function (NP_011719); ACR2_cerevisiae, S. cerevisiae arsenate reductase (NP_015526); IBP1_pombe, Sch. pombe small CDC25-like protein (AL096796); Orysa;CDC25;1, O. sativa protein of unknown function (NP_922597); Arath;CDC25_Arabidop, this work (NP_568119).
Fig. 2.
Activation of the A. thaliana CDK kinase activity by the Arath;CDC25 protein. The Arabidopsis CDK complexes were partially purified on p10CKS1At beads. The CDK-associated kinase activity was determined in the absence (lane 1) or presence of 4 (lane 2), 8 (lane 3), and 20 μg (lane 4) of Arath;CDC25 protein and 8 μg of Arath;CDC25 protein and 40 nM specific CDC25 inhibitor NSC95397 (lane 5). The graph illustrates the quantification of kinase activity by phosphorimager detection of the incorporation of radioactive phosphate by histone H1 normalized to the CDKA amount, control by Western blot with anti-PSTAIRE antibodies. Numbers on y axis are cpm × 10-3.
Fig. 3.
Ribbon representation in the same orientation of the backbone of a representative conformer of the Arath;CDC25 protein (Left) and the human CDC25A (Right) (PDB entry 1C25, ref. 4). The conserved five-stranded β-sheet and α4 helices of the structures have been superposed as described in Results. The catalytic loop is indicated by the HC(X)5R motif. The zinc ion is represented as a black sphere.
Fig. 4.
Map of the chemical-shift perturbation data on a surface and a ribbon representation in the same orientation of the Arath;CDC25 protein. Residues are colored according to the chemical-shift perturbation observed upon addition of an excess of pTyr15CDKA (Upper) or pThr14pTyr15CDKA (Lower) peptide. The color scale is as follows: yellow <0.04 ppm, orange between 0.04 and 0.1 ppm, and red >0.1 ppm combined 1H-15N chemical-shift difference between the free and the peptide-bound form of the Arath;CDC25 protein. The zinc ion is not represented. On the left of the structure representations, overlaid sections of 1HN-15N-HSQC spectra acquired on Arath;CDC25 before (in red contours) and after (in blue contours) addition of 20 molar equivalent of pTyr15CDKA and pThr14pTyr15CDKA peptides.
Similar articles
- Characterization of the Arabidopsis thaliana Arath;CDC25 dual-specificity tyrosine phosphatase.
Landrieu I, Hassan S, Sauty M, Dewitte F, Wieruszeski JM, Inzé D, De Veylder L, Lippens G. Landrieu I, et al. Biochem Biophys Res Commun. 2004 Sep 24;322(3):734-9. doi: 10.1016/j.bbrc.2004.07.182. Biochem Biophys Res Commun. 2004. PMID: 15336525 - A model of Cdc25 phosphatase catalytic domain and Cdk-interaction surface based on the presence of a rhodanese homology domain.
Hofmann K, Bucher P, Kajava AV. Hofmann K, et al. J Mol Biol. 1998 Sep 11;282(1):195-208. doi: 10.1006/jmbi.1998.1998. J Mol Biol. 1998. PMID: 9733650 - Arabidopsis T-DNA insertional lines for CDC25 are hypersensitive to hydroxyurea but not to zeocin or salt stress.
Spadafora ND, Doonan JH, Herbert RJ, Bitonti MB, Wallace E, Rogers HJ, Francis D. Spadafora ND, et al. Ann Bot. 2011 May;107(7):1183-92. doi: 10.1093/aob/mcq142. Epub 2010 Jul 20. Ann Bot. 2011. PMID: 20647223 Free PMC article. - The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations.
Bordo D, Bork P. Bordo D, et al. EMBO Rep. 2002 Aug;3(8):741-6. doi: 10.1093/embo-reports/kvf150. EMBO Rep. 2002. PMID: 12151332 Free PMC article. Review. - Potential of CDC25 phosphatases in cancer research and treatment: key to precision medicine.
Dakilah I, Harb A, Abu-Gharbieh E, El-Huneidi W, Taneera J, Hamoudi R, Semreen MH, Bustanji Y. Dakilah I, et al. Front Pharmacol. 2024 Jan 19;15:1324001. doi: 10.3389/fphar.2024.1324001. eCollection 2024. Front Pharmacol. 2024. PMID: 38313315 Free PMC article. Review.
Cited by
- Regulatory dephosphorylation of CDK at G₂/M in plants: yeast mitotic phosphatase cdc25 induces cytokinin-like effects in transgenic tobacco morphogenesis.
Lipavská H, Masková P, Vojvodová P. Lipavská H, et al. Ann Bot. 2011 May;107(7):1071-86. doi: 10.1093/aob/mcr016. Epub 2011 Feb 20. Ann Bot. 2011. PMID: 21339187 Free PMC article. - A vacuolar arsenite transporter necessary for arsenic tolerance in the arsenic hyperaccumulating fern Pteris vittata is missing in flowering plants.
Indriolo E, Na G, Ellis D, Salt DE, Banks JA. Indriolo E, et al. Plant Cell. 2010 Jun;22(6):2045-57. doi: 10.1105/tpc.109.069773. Epub 2010 Jun 8. Plant Cell. 2010. PMID: 20530755 Free PMC article. - Identification of conserved tyrosine residues important for gibberellin sensitivity of Arabidopsis RGL2 protein.
Hussain A, Cao D, Peng J. Hussain A, et al. Planta. 2007 Jul;226(2):475-83. doi: 10.1007/s00425-007-0497-z. Epub 2007 Mar 1. Planta. 2007. PMID: 17333251 - Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation.
Pina C, Pinto F, Feijó JA, Becker JD. Pina C, et al. Plant Physiol. 2005 Jun;138(2):744-56. doi: 10.1104/pp.104.057935. Epub 2005 May 20. Plant Physiol. 2005. PMID: 15908605 Free PMC article. - Protein structure database search and evolutionary classification.
Yang JM, Tung CH. Yang JM, et al. Nucleic Acids Res. 2006 Aug 2;34(13):3646-59. doi: 10.1093/nar/gkl395. Print 2006. Nucleic Acids Res. 2006. PMID: 16885238 Free PMC article.
References
- Dunphy, W. G. & Kumagai, A. (1991) Cell 67, 189-196. - PubMed
- Kumagai, A. & Dunphy, W. G. (1991) Cell 64, 903-914. - PubMed
- Nilsson, I. & Hoffmann, I. (2000) Prog. Cell Cycle Res. 4, 107-114. - PubMed
- Fauman, E. B., Cogswell, J. P., Lovejoy, B., Rocque, W. J., Holmes, W., Montana, V. G., Piwnica-Worms, H., Rink, M. J. & Saper, M. A. (1998) Cell 93, 617-625. - PubMed
- Hofmann, K., Bucher, P. & Kajava, A. V. (1998) J. Mol. Biol. 282, 195-208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials