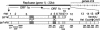Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins - PubMed (original) (raw)
Comparative Study
Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins
Erik Prentice et al. J Virol. 2004 Sep.
Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) encodes proteins required for RNA transcription and genome replication as large polyproteins that are proteolytically processed by virus-encoded proteinases to produce mature replicase proteins. In this report, we generated antibodies against SARS-CoV predicted replicase protein and used the antibodies to identify and characterize 12 of the 16 predicted mature replicase proteins (nsp1, nsp2, nsp3, nsp4, nsp5, nsp8, nsp9, nsp12, nsp13, nsp14, nsp15, and nsp16) in SARS-CoV-infected Vero cells. Immunoblot analysis of infected-cell lysates identified proteins of the predicted sizes. Immunofluorescence microscopy detected similar patterns of punctate perinuclear and distributed cytoplasmic foci with all replicase antibodies and as early as 6 h postinfection. Dual-labeling studies demonstrated colocalization of replicase protein nsp8 with nsp2 and nsp3 in cytoplasmic complexes and also with LC3, a protein marker for autophagic vacuoles. Antibodies directed against mouse hepatitis virus (MHV) virions and against the putative RNA-dependent RNA polymerase (Pol) detected SARS-CoV nucleocapsid and nsp12 (Pol), respectively, in SARS-CoV-infected Vero cells. These results confirm the predicted protein processing pattern for mature SARS-CoV replicase proteins, demonstrate localization of replicase proteins to cytoplasmic complexes containing markers for autophagosome membranes, and suggest conservation of protein epitopes in the replicase and nucleocapsid of SARS-CoV and the group II coronavirus, MHV. Further, the results demonstrate the ability of replicase antibodies to detect SARS-CoV-infected cells as early as 6 h postinfection and thus represent important tools for studies of SARS-CoV replication, inhibition, and diagnosis.
Copyright 2004 American Society for Microbiology
Figures
FIG. 1.
Predicted SARS-CoV replicase gene organization, mature proteins, and antibodies. The location of the SARS-CoV replicase gene in the genome is depicted, with ORF1a and ORF1b shown. The protein domains of polyprotein 1a (pp1a) and the frameshift-fusion polyprotein 1ab (pp1a/b) are shown. Protein domains are indicated by vertical bars, by nonstructural protein number (nsp), and by activities or putative or known functions. The processing of the replicase polyprotein is mediated by two proteinases, the PLP and the 3CL-pro. Black rectangles below proteins indicate regions cloned and expressed as proteins for induction of antibodies (numbers 231 to 265).
FIG. 2.
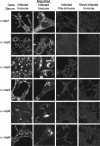
Identification of ORF1a SARS-CoV replicase proteins by immunoblotting. Lysates of SARS-CoV-infected (I) and mock-infected (M) Vero cells were separated by SDS-PAGE, transferred to nitrocellulose, and probed with the anti-nsp1 (α-nsp1), -nsp2, -nsp3, -nsp4, -nsp5, -nsp8 or -nsp9 antibody. Additionally, preimmune (P) sera from the same rabbits were used to immunoblot infected-cell lysates. Marker proteins are to the left of the blots, and observed masses of specific products are to the right of the blots.
FIG. 3.
Localization of SARS-CoV ORF1a replicase proteins in Vero cells. Vero cells on glass coverslips were infected with SARS-CoV or were mock infected for 12 h prior to fixation with methanol. Cells were incubated with the indicated antibodies (e.g., anti-nsp1 [α-nsp1]) and a secondary Alexa 488 and imaged by indirect fluorescent-antibody assay, using a Zeiss 510 confocal microscope as described in Materials and Methods. Images in the second column were obtained at higher magnification to show single-cell details of fluorescence labeling.
FIG. 4.
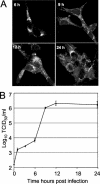
(A) Time course of nsp1 detection. Vero cells were infected with SARS-CoV, fixed at 3, 6, 9, 12, and 24 h p.i., and labeled with anti-nsp1 antibodies. No specific signal was observed at 3 h p.i. (not shown). (B) Single-cycle growth of SARS-CoV growth in Vero cells. Vero cell monolayers were infected with SARS-CoV at an MOI of one TCID50/cell, and samples of medium were obtained at times indicated postinfection and assayed by TCID50. Error bars indicate standard deviations of four measurements in each of two independent experiments.
FIG. 5.
Identification of ORF1b SARS-CoV replicase proteins by immunoblotting and immunofluorescence. (A) Lysates of SARS-CoV-infected (I) and mock-infected (M) cells were separated by SDS-PAGE, transferred to nitrocellulose, and probed with the anti-nsp12 (α-nsp12), -nsp13, -nsp14, -nsp15, or -nsp16 antibody. Additionally, preimmune (P) sera were used to probe blots. Mass markers are to the left of the blots, and observed masses of specific proteins are to the right of the blots. (B) Vero cells were infected with SARS-CoV or were mock infected for 12 h prior to fixation with methanol. Cells were labeled with the anti-nsp13 antibody and imaged as described in Materials and Methods. The second column is the image obtained at increased magnification to show detail of the fluorescence image.
FIG. 6.
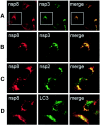
Colocalization of replicase proteins and cellular proteins in SARS-CoV-infected Vero Cells. Vero cells on glass coverslips were infected with SARS-CoV (Urbani strain) at an MOI of 1.0 TCID50 per cell. At 6 h p.i., cells were fixed with 100% methanol and processed for dual labeling using guinea pig anti-nsp8 (red, Alexa 546 in all panels) and rabbit anti-nsp3, anti-nsp 2, or anti-LC3 (green, Alexa 488). (A) Colocalization of nsp8 (red) and nsp3 (green). (B) Magnification of panel A cells (white outline) to show single-cell detail. (C) Colocalization of nsp8 (red) and nsp2 (green). (D) Colocalization of nsp8 (red) and LC3 (green). See the text for details. The merged column shows colocalization of red and green signal as yellow pixels. Images were obtained on a Zeiss LSM 510 confocal microscope.
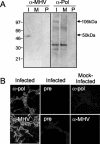
FIG. 7.
MHV antibodies detect SARS-CoV proteins. (A) Identification of replicase proteins by immunoblotting. SARS-CoV-infected (I) and mock-infected (M) cell lysates were separated by SDS-PAGE, transferred to nitrocellulose, and labeled with anti-MHV (α-MHV) or MHV anti-Pol (α-Pol) antibody. Additionally, preimmune (P) sera were used to label blots. Mass markers are to the left, and masses of specific proteins are given to the right of the blots. (B) Antibodies against MHV-Pol and MHV were used to label SARS-CoV-infected or mock-infected Vero cells by immunofluorescence. Infected Vero cells were stained with immune sera. Fluorescence was not detected in mock-infected cells with immune sera or in infected cells stained with preimmune sera.
Similar articles
- Expression Profile and Localization of SARS-CoV-2 Nonstructural Replicase Proteins in Infected Cells.
Shi FS, Yu Y, Li YL, Cui L, Zhao Z, Wang M, Wang B, Zhang R, Huang YW. Shi FS, et al. Microbiol Spectr. 2022 Aug 31;10(4):e0074422. doi: 10.1128/spectrum.00744-22. Epub 2022 Jun 22. Microbiol Spectr. 2022. PMID: 35730969 Free PMC article. - Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity.
Harcourt BH, Jukneliene D, Kanjanahaluethai A, Bechill J, Severson KM, Smith CM, Rota PA, Baker SC. Harcourt BH, et al. J Virol. 2004 Dec;78(24):13600-12. doi: 10.1128/JVI.78.24.13600-13612.2004. J Virol. 2004. PMID: 15564471 Free PMC article. - The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication.
Graham RL, Sims AC, Brockway SM, Baric RS, Denison MR. Graham RL, et al. J Virol. 2005 Nov;79(21):13399-411. doi: 10.1128/JVI.79.21.13399-13411.2005. J Virol. 2005. PMID: 16227261 Free PMC article. - Characterization of viral proteins encoded by the SARS-coronavirus genome.
Tan YJ, Lim SG, Hong W. Tan YJ, et al. Antiviral Res. 2005 Feb;65(2):69-78. doi: 10.1016/j.antiviral.2004.10.001. Antiviral Res. 2005. PMID: 15708633 Free PMC article. Review. - Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus.
Tan YJ, Lim SG, Hong W. Tan YJ, et al. Antiviral Res. 2006 Nov;72(2):78-88. doi: 10.1016/j.antiviral.2006.05.010. Epub 2006 Jun 6. Antiviral Res. 2006. PMID: 16820226 Free PMC article. Review.
Cited by
- Fluvoxamine: A Review of Its Mechanism of Action and Its Role in COVID-19.
Sukhatme VP, Reiersen AM, Vayttaden SJ, Sukhatme VV. Sukhatme VP, et al. Front Pharmacol. 2021 Apr 20;12:652688. doi: 10.3389/fphar.2021.652688. eCollection 2021. Front Pharmacol. 2021. PMID: 33959018 Free PMC article. - SARS-CoV-2 Dissemination Through Peripheral Nerves Explains Multiple Organ Injury.
Fenrich M, Mrdenovic S, Balog M, Tomic S, Zjalic M, Roncevic A, Mandic D, Debeljak Z, Heffer M. Fenrich M, et al. Front Cell Neurosci. 2020 Aug 5;14:229. doi: 10.3389/fncel.2020.00229. eCollection 2020. Front Cell Neurosci. 2020. PMID: 32848621 Free PMC article. - Characterization of a novel bat-HKU2-like swine enteric alphacoronavirus (SeACoV) infection in cultured cells and development of a SeACoV infectious clone.
Yang YL, Liang QZ, Xu SY, Mazing E, Xu GH, Peng L, Qin P, Wang B, Huang YW. Yang YL, et al. Virology. 2019 Oct;536:110-118. doi: 10.1016/j.virol.2019.08.006. Epub 2019 Aug 9. Virology. 2019. PMID: 31419711 Free PMC article. - Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs.
Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Brooun A, Griffith M, Moy K, Yadav MK, Velasquez J, Buchmeier MJ, Stevens RC, Kuhn P. Joseph JS, et al. J Virol. 2006 Aug;80(16):7894-901. doi: 10.1128/JVI.00467-06. J Virol. 2006. PMID: 16873246 Free PMC article. - Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV.
Brierley I, Dos Ramos FJ. Brierley I, et al. Virus Res. 2006 Jul;119(1):29-42. doi: 10.1016/j.virusres.2005.10.008. Epub 2005 Nov 28. Virus Res. 2006. PMID: 16310880 Free PMC article. Review.
References
- Bi, W., J. D. Pinon, S. Hughes, P. J. Bonilla, K. V. Holmes, S. R. Weiss, and J. L. Leibowitz. 1998. Localization of mouse hepatitis virus open reading frame 1a derived proteins. J. Neurovirol. 4:594-605. - PubMed
- Booth, C. M., L. M. Matukas, G. A. Tomlinson, A. R. Rachlis, D. B. Rose, H. A. Dwosh, S. L. Walmsley, T. Mazzulli, M. Avendano, P. Derkach, I. E. Ephtimios, I. Kitai, B. D. Mederski, S. B. Shadowitz, W. L. Gold, L. A. Hawryluck, E. Rea, J. S. Chenkin, D. W. Cescon, S. M. Poutanen, and A. S. Detsky. 2003. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA 289:2801-2809. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HL007751/HL/NHLBI NIH HHS/United States
- 5 T32 HL07751/HL/NHLBI NIH HHS/United States
- R01 AI050083/AI/NIAID NIH HHS/United States
- AI50083-S1/AI/NIAID NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- CA68485/CA/NCI NIH HHS/United States
- DK20593/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous