A cotranscriptional model for 3'-end processing of the Saccharomyces cerevisiae pre-ribosomal RNA precursor - PubMed (original) (raw)
A cotranscriptional model for 3'-end processing of the Saccharomyces cerevisiae pre-ribosomal RNA precursor
Anthony K Henras et al. RNA. 2004 Oct.
Abstract
Cleavage of the Saccharomyces cerevisiae primary ribosomal RNA (rRNA) transcript in the 3' external transcribed spacer (ETS) by Rnt1p generates the 35S pre-rRNA, the earliest detectable species in the pre-rRNA processing pathway. In this study we show that Rnt1p is concentrated in a subnucleolar dot-shaped territory distinct from the nucleolar body. The 35S pre-rRNA is localized at the periphery of the Rnt1p dot, in a pattern that suggests a diffusion of the 35S pre-rRNA from the site of Rnt1p processing. When plasmid-borne versions of the rDNA are used to express rRNAs, the Rnt1p territory reorganizes around these plasmids, suggesting a close association between Rnt1p and the plasmid-borne rDNA units. Rnt1p was found associated with the endogenous rDNA by chromatin immunoprecipitation. Deletion of functionally important Rnt1p domains result in a loss of the dot-shaped territory, showing that this subnucleolar territory corresponds to a functional site of processing. These results show that a large fraction of Rnt1p is localized at the site of transcription of the rDNA, suggesting that the cleavage of the primary pre-rRNA transcript to generate the 35S pre-rRNA is a cotranscriptional event.
Copyright 2004 RNA Society
Figures
FIGURE 1.
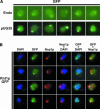
Rnt1p is localized in the nucleolus and in the nucleoplasm. (A) Subcellular localization of Rnt1p–GFP expressed from the endogenous locus (top row, endo) and from the pUG35 plasmid (pUG35). (B) Subcellular localization of Rnt1pGFP expressed from the pUG35 vector. Shown are pictures obtained from the haploid strain rnt1 :: TRP transformed with plasmids pUG35–RNT1, and treated for immunofluorescence with anti-Nop1p antibodies. From left to right, DAPI staining (blue), Rnt1p fused to the GFP (green), endogenous Nop1p (red), and merged images enabling the simultaneous visualization of both Nop1p and DAPI, GFP and DAPI, and Nop1p and GFP. (C) The nucleolar structure containing Rnt1p and the NB correspond to different nucleolar subdomains. (Left) Localization of Rnt1p–GFP (green); (middle) localization of the artificial snoRNA in the NB obtained by FISH (red); (right) merged image to which the DAPI staining has been added. (D) Rnt1p does not colocalize with the U14 snoRNA. (Left) Localization of Rnt1p–GFP (green); (middle) localization of the U14 snoRNA obtained by FISH (red); (right) merged image.
FIGURE 1.
Rnt1p is localized in the nucleolus and in the nucleoplasm. (A) Subcellular localization of Rnt1p–GFP expressed from the endogenous locus (top row, endo) and from the pUG35 plasmid (pUG35). (B) Subcellular localization of Rnt1pGFP expressed from the pUG35 vector. Shown are pictures obtained from the haploid strain rnt1 :: TRP transformed with plasmids pUG35–RNT1, and treated for immunofluorescence with anti-Nop1p antibodies. From left to right, DAPI staining (blue), Rnt1p fused to the GFP (green), endogenous Nop1p (red), and merged images enabling the simultaneous visualization of both Nop1p and DAPI, GFP and DAPI, and Nop1p and GFP. (C) The nucleolar structure containing Rnt1p and the NB correspond to different nucleolar subdomains. (Left) Localization of Rnt1p–GFP (green); (middle) localization of the artificial snoRNA in the NB obtained by FISH (red); (right) merged image to which the DAPI staining has been added. (D) Rnt1p does not colocalize with the U14 snoRNA. (Left) Localization of Rnt1p–GFP (green); (middle) localization of the U14 snoRNA obtained by FISH (red); (right) merged image.
FIGURE 2.
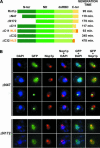
Localization of Rnt1p with the 35S pre-rRNA precursor and rDNA. (A) The 35S pre-rRNA is localized at the immediate periphery of the nucleolar dot containing Rnt1p. Shown is a schematic representation of the rRNA primary transcript. The red line indicates the site of hybridization of the oligonucleotide used in the FISH experiment. The hairpin indicates the site of Rnt1p cleavage. Shown are pictures obtained from the haploid rnt1::TRP strain transformed with plasmids pUG35–RNT1, and treated for pre-rRNA FISH. (Left) Rnt1p–GFP (green); (middle) localization of the 35S pre-rRNA (red); (right) merged image. (B) Rnt1p accumulates within several nuclear foci in a yeast mutant strain expressing the rRNAs from multicopy plasmids. Shown are (immuno)fluorescence pictures obtained from strain NOY758 transformed with pUG35–RNT1. Legends as in Figure 1A ▶. (C) Rnt1p strongly accumulates within several nuclear foci in a yeast mutant strain expressing the rRNAs from multicopy plasmids containing an RNA polymerase II promoter. Shown are (immuno)fluorescence pictures obtained from strain NOY759 transformed with pUG35–RNT1. Legends as in Figure 1A ▶.
FIGURE 3.
Rnt1p is associated with the rDNA chromatin. (A) Organization of the rDNA repeat and localization of the PCR products obtained in the ChIP procedure. (B) Shown are α32P-labeled PCR products obtained from Input DNA (lane 1) or DNA extracted from immunoprecipitation reactions in extracts prepared from the rnt1:TRP, pUG35–RNT1 strain using anti-GFP IgGs (αGFP, lane 2), irrelevant anti-FLAG IgGs (α-FLAG, lane 3), or with Protein G-conjugated sepharose beads only (Beads, lane 4), using oligonucleotide pairs hybridizing to the indicated DNA sequences. (C) Quantification of the ChIP data. Five independent ChIP experiments were carried out as described in Materials and Methods. For each experiment, the intensity of the radioactive bands was measured using Image Quant. The relative immunoprecipitation efficiency of the different DNA targets by the anti-GFP antibodies was calculated as follows: For a given primer pair, the average intensity of the bands obtained in the negative control experiments and reflecting the immunoprecipitation background (“α-FLAG” or “Beads only” controls) was subtracted from the intensity of the band obtained with the relevant, α-GFP antibodies. The resulting value was then divided by the intensity of the band corresponding to the INPUT experiment. This value was then divided by the value obtained for the CUP1 signal (negative control). Error bars indicate the standard deviation. Because CUP1 was used as an internal standard for each experiment, no error bar is included.
FIGURE 4.
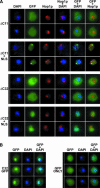
The N-terminal domain of Rnt1p is not required for the import of Rnt1p into the nucleus of yeast cells. (A) Schematic representation of the constructs used in the truncation analysis. Shown are the boundaries of the Rnt1p domains and of the truncations generated, and the generation time of the the rnt1 :: TRP transformed with the pUG35 derivatives expressing the different truncated versions of the protein. The generation time of rnt1 :: TRP transformed with plasmid pUG35 (knockout control) is 440 min. (N-ter) N-terminal domain, (ND) RNase III Nuclease Domain, (dsRBD) double-stranded RNA binding domain, (C-ter) C-terminal domain. (B) Shown are (immuno)fluorescence images obtained with the haploid strain rnt1 :: TRP transformed with plasmids pUG35–RNT1ΔN47 or pUG35–RNT1ΔN172. Legends as in Figure 1A ▶
FIGURE 5.
The C-terminal domain of Rnt1p is necessary and sufficient for nuclear import. (A) Deletion of the C-terminal amino acids of Rnt1p perturbs nuclear import. Shown are (immuno)fluorescence images obtained with the haploid strain rnt1 :: TRP transformed with plasmids pUG35–RNT1ΔC11, pUG35–RNT1ΔC11NLS, pUG35–RNT1ΔC32, or pUG35–RNT1ΔC32NLS. Legends as in Figure 1A ▶.(B) The C-terminal domain of Rnt1p is a bona fide nuclear targeting signal. Shown are fluorescence pictures obtained from the haploid strain BMA64 (wild type) transformed with plasmid pUG35-C32 or pUG35.
Similar articles
- The role of the 3' external transcribed spacer in yeast pre-rRNA processing.
Allmang C, Tollervey D. Allmang C, et al. J Mol Biol. 1998 Apr 24;278(1):67-78. doi: 10.1006/jmbi.1998.1693. J Mol Biol. 1998. PMID: 9571034 - Genome-wide prediction and analysis of yeast RNase III-dependent snoRNA processing signals.
Ghazal G, Ge D, Gervais-Bird J, Gagnon J, Abou Elela S. Ghazal G, et al. Mol Cell Biol. 2005 Apr;25(8):2981-94. doi: 10.1128/MCB.25.8.2981-2994.2005. Mol Cell Biol. 2005. PMID: 15798187 Free PMC article. - The RNA catabolic enzymes Rex4p, Rnt1p, and Dbr1p show genetic interaction with trans-acting factors involved in processing of ITS1 in Saccharomyces cerevisiae pre-rRNA.
Faber AW, Vos JC, Vos HR, Ghazal G, Elela SA, Raué HA. Faber AW, et al. RNA. 2004 Dec;10(12):1946-56. doi: 10.1261/rna.7155904. Epub 2004 Nov 3. RNA. 2004. PMID: 15525710 Free PMC article. - Structure and function of Rnt1p: An alternative to RNAi for targeted RNA degradation.
Abou Elela S, Ji X. Abou Elela S, et al. Wiley Interdiscip Rev RNA. 2019 May;10(3):e1521. doi: 10.1002/wrna.1521. Epub 2018 Dec 11. Wiley Interdiscip Rev RNA. 2019. PMID: 30548404 Review. - DNA protein interactions at the rRNA of Saccharomyces cerevisiae.
Cioci F, Di Felice F, Chiani F, Camilloni G. Cioci F, et al. Ital J Biochem. 2007 Jun;56(2):81-90. Ital J Biochem. 2007. PMID: 17722648 Review.
Cited by
- Structure of a yeast RNase III dsRBD complex with a noncanonical RNA substrate provides new insights into binding specificity of dsRBDs.
Wang Z, Hartman E, Roy K, Chanfreau G, Feigon J. Wang Z, et al. Structure. 2011 Jul 13;19(7):999-1010. doi: 10.1016/j.str.2011.03.022. Structure. 2011. PMID: 21742266 Free PMC article. - Nuclear fate of yeast snoRNA is determined by co-transcriptional Rnt1 cleavage.
Grzechnik P, Szczepaniak SA, Dhir S, Pastucha A, Parslow H, Matuszek Z, Mischo HE, Kufel J, Proudfoot NJ. Grzechnik P, et al. Nat Commun. 2018 May 3;9(1):1783. doi: 10.1038/s41467-018-04094-y. Nat Commun. 2018. PMID: 29725044 Free PMC article. - IT'S 2 for the price of 1: Multifaceted ITS2 processing machines in RNA and DNA maintenance.
Pillon MC, Lo YH, Stanley RE. Pillon MC, et al. DNA Repair (Amst). 2019 Sep;81:102653. doi: 10.1016/j.dnarep.2019.102653. Epub 2019 Jul 8. DNA Repair (Amst). 2019. PMID: 31324529 Free PMC article. Review. - Efficient termination of transcription by RNA polymerase I requires the 5' exonuclease Rat1 in yeast.
El Hage A, Koper M, Kufel J, Tollervey D. El Hage A, et al. Genes Dev. 2008 Apr 15;22(8):1069-81. doi: 10.1101/gad.463708. Genes Dev. 2008. PMID: 18413717 Free PMC article. - Transcription termination by nuclear RNA polymerases.
Richard P, Manley JL. Richard P, et al. Genes Dev. 2009 Jun 1;23(11):1247-69. doi: 10.1101/gad.1792809. Genes Dev. 2009. PMID: 19487567 Free PMC article.
References
- Abou Elela, S., Igel, H., and Ares Jr., M. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85: 115–124. - PubMed
- Allmang, C. and Tollervey, D. 1998. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 278: 67–78. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases