A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products - PubMed (original) (raw)
A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products
Kendy K Y Wong et al. J Bacteriol. 2004 Sep.
Abstract
The PrfA protein of Listeria monocytogenes functions as a key regulatory factor for the coordinated expression of many virulence genes during bacterial infection of host cells. PrfA activity is controlled by multiple regulatory mechanisms, including an apparent requirement for either the presence of a cofactor or some form of posttranslational modification that regulates the activation of PrfA. In this study, we describe the identification and characterization of a novel PrfA mutation that results in constitutive activation of the PrfA protein. The PrfA L140F mutation was found to confer high-level expression of PrfA-regulated genes and to be functionally dominant over the wild-type allele. The presence of the PrfA L140F mutation resulted in the aggregation of L. monocytogenes in broth culture and, unlike previously described prfA mutations, appeared to be slightly toxic to the bacteria. High-level PrfA-dependent gene expression showed no additional increase in L. monocytogenes strains containing an additional copy of prfA L140F despite a >4-fold increase in PrfA protein levels. In contrast, the introduction of multiple copies of the wild-type prfA allele to L. monocytogenes resulted in a corresponding increase in PrfA-dependent gene expression, although overall expression levels remained far below those observed for PrfA L140F strains. These results suggest a hierarchy of PrfA regulation, such that the relative levels of PrfA protein present within the cell correlate with the levels of PrfA-dependent gene expression when the protein is not in its fully activated state; however, saturating levels of the protein are then quickly reached when PrfA is converted to its active form. Regulation of the PrfA activation status must be an important facet of L. monocytogenes survival, as mutations that result in constitutive PrfA activation may have deleterious consequences for bacterial physiology.
Figures
FIG. 1.
Locations of PrfA mutations with respect to predicted protein secondary-structure motifs and functional regions. The previously described PrfA* mutation (G145S) is included for comparison (37). AR, activation region that may form contacts with RNA polymerase; Leu-zip, leucine zipper-like motif. Depictions of structural motifs and functional regions are adapted from Goebel et al. (17).
FIG. 2.
Overnight cultures of L. monocytogenes strains grown in BHI broth at 37°C without shaking. The strain numbers and relevant genotypes are shown underneath the respective culture tubes. 476, NF-L476 (WT); 879, NF-L879 (EMS L140F mutant); 1041, NF-L1041 (Δ_prfA_ + WT_i_); 1011, NF-L1011 (Δ_prfA_ + L140F_i_); 1067, NF-L1067 (L140F + L140F_i_); 1006, NF-L1006 (WT + pPL2_i_); 1008, NF-L1008 (WT + L140F_i_).
FIG. 3.
Western analysis of PrfA protein levels produced by the various L. monocytogenes strains. Soluble bacterial whole-cell lysates were prepared from mid-log-phase cultures. The strain numbers and relevant genotypes are shown above the lanes. PrfA was detected by using a monoclonal antibody against PrfA and an alkaline phosphatase-conjugated goat anti-mouse secondary antibody. (A) PrfA protein levels from L. monocytogenes integrant strains in the NF-L1003 (Δ_prfA_) background. Equal amounts of total protein (23 μg) solubilized in SDS-PAGE sample buffer were loaded for each sample. (B) Quantitative comparison of PrfA protein levels from WT strain NF-L476 (476) and original EMS mutant strain NF-L879 (879), with the amount of total proteins loaded in each lane indicated. (C) Quantitative comparison of PrfA protein levels from integrant strains NF-L1041 (Δ_prfA_ + WT_i_) (1041) and NF-L1011 (Δ_prfA_ + L140F_i_) (1011), with the amount of total proteins loaded in each lane indicated.
FIG. 4.
actA expression of PrfA mutant strains and corresponding control strains during growth in broth culture. Units of GUS activity were determined at the indicated time intervals as described in Materials and Methods and were normalized for bacterial culture OD595 (51), using 4-methylumbilliferyl-β-
d
-glucuronide as the substrate. The data shown are from duplicate samples and are representative of at least three independent experiments, expressed as average ± standard error. (A) Growth of WT and PrfA mutant strains in BHI broth at 37°C as measured by OD595 of cultures. (B) actA expression from strains NF-L476 (WT), NF-L1003 (Δ_prfA_), NF-L1009 (Δ_prfA_ + pPL2_i_), NF-L1041 (Δ_prfA_ + WT_i_), and NF-L1042 (Δ_prfA_ + L147P_i_). (C) actA expression from strains NF-L476 (WT), NF-L879 (EMS L140F), and NF-L1011 (Δ_prfA_ + L140F_i_).
FIG. 5.
PlcB (lecithinase) phenotypes of L. monocytogenes mutants and corresponding control strains on BHI medium overlaid with molten agar containing activated-charcoal-treated (0.2% [wt/vol]) BHI and 5% (vol/vol) 1:1 egg yolk-PBS solution. The plates were incubated at 37°C overnight. 476, NF-L476 (WT); 879, NF-L879 (EMS L140F mutant); 1003, NF-L1003 (Δ_prfA_); 1006, NF-L1006 (WT + pPL2_i_); 1039, NF-L1039 (WT + WT_i_); 1040, NF-L1040 (WT + L147P_i_); 1008, NF-L1008 (WT + L140F_i_); 1009, NF-L1009 (Δ_prfA_ + pPL2_i_); 1041, NF-L1041 (Δ_prfA_ + WT_i_); 1042, NF-L1042 (Δ_prfA_ + L147P_i_); 1011, NF-L1011 (Δ_prfA_ + L140F_i_); 1067, NF-L1067 (L140F + L140F_i_).
FIG. 6.
Western analysis of PrfA protein levels produced by the various L. monocytogenes strains. Soluble bacterial whole-cell lysates were prepared from mid-log-phase cultures. The strain numbers and relevant genotypes are shown above the lanes. PrfA was detected by using a monoclonal antibody against PrfA and an alkaline phosphatase-conjugated goat anti-mouse secondary antibody. (A) PrfA protein levels from L. monocytogenes integrant strains in the WT strain NF-L476 background. Equal amounts of total protein (23 μg) solubilized in SDS-PAGE sample buffer were loaded for each sample. (B) Quantitative comparison of PrfA protein levels from integrant strains NF-L1006 (WT + pPL2_i_) (1006) and NF-L1008 (WT + L140F_i_) (1008). (C) Quantitative comparison of PrfA protein levels from integrant strains NF-L1006 (WT + pPL2_i_) and NF-L1039 (WT + WT_i_) (1039). (D) Quantitative comparison of PrfA protein levels from EMS mutant strain NF-L879 (879), NF-L1003 (Δ_prfA_) (1003), and integrant strain NF-L1067 (L140F + L140F_i_) (1067). The amounts of total proteins loaded in each lane in panels B, C, and D are indicated.
FIG. 7.
actA expression of PrfA mutant strains and corresponding control strains during growth in broth culture. Units of GUS activity were determined at the indicated time intervals as described in Materials and Methods and were normalized for bacterial culture OD595 (51) using 4-methylumbilliferyl-β-
d
-glucuronide as the substrate. The data shown are from duplicate samples and are representative of at least three independent experiments, expressed as average ± standarderror. (A) Growth of WT and PrfA mutant strains in BHI broth at 37°C as measured by OD595 of cultures. (B) actA expression from strains NF-L476 (WT), NF-L1003 (Δ_prfA_), NF-L1006 (WT + pPL2_i_), NF-L1039 (WT + WT_i_), and NF-L1040 (WT + L147P_i_). (C) actA expression from strains NF-L476 (WT), NF-L879 (EMS L140F), and NF-L1008 (WT + L140F_i_). (D) actA expression from strains NF-L476 (WT), NF-L879 (EMS L140F), and NF-L1067 (L140F + L140F_i_).
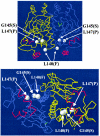
FIG. 8.
Locations of PrfA mutations in the crystal structure of PrfA (
). The two diagrams illustrate two different views of the PrfA dimer. The blue and yellow traces are the Cα traces of the two monomers of PrfA. The HTH motifs are shown in red. The previously described PrfA* mutation (G145S) is included for comparison (37).
Similar articles
- A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes.
Ripio MT, Domínguez-Bernal G, Lara M, Suárez M, Vazquez-Boland JA. Ripio MT, et al. J Bacteriol. 1997 Mar;179(5):1533-40. doi: 10.1128/jb.179.5.1533-1540.1997. J Bacteriol. 1997. PMID: 9045810 Free PMC article. - Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products.
Shetron-Rama LM, Mueller K, Bravo JM, Bouwer HG, Way SS, Freitag NE. Shetron-Rama LM, et al. Mol Microbiol. 2003 Jun;48(6):1537-51. doi: 10.1046/j.1365-2958.2003.03534.x. Mol Microbiol. 2003. PMID: 12791137 - New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA.
Vega Y, Rauch M, Banfield MJ, Ermolaeva S, Scortti M, Goebel W, Vázquez-Boland JA. Vega Y, et al. Mol Microbiol. 2004 Jun;52(6):1553-65. doi: 10.1111/j.1365-2958.2004.04052.x. Mol Microbiol. 2004. PMID: 15186408 - Regulation of virulence genes in Listeria.
Kreft J, Vázquez-Boland JA. Kreft J, et al. Int J Med Microbiol. 2001 May;291(2):145-57. doi: 10.1078/1438-4221-00111. Int J Med Microbiol. 2001. PMID: 11437337 Review. - From hot dogs to host cells: how the bacterial pathogen Listeria monocytogenes regulates virulence gene expression.
Freitag NE. Freitag NE. Future Microbiol. 2006 Jun;1(1):89-101. doi: 10.2217/17460913.1.1.89. Future Microbiol. 2006. PMID: 17661688 Review.
Cited by
- Regulation of Listeria monocytogenes Virulence.
Johansson J, Freitag NE. Johansson J, et al. Microbiol Spectr. 2019 Jul;7(4):10.1128/microbiolspec.gpp3-0064-2019. doi: 10.1128/microbiolspec.GPP3-0064-2019. Microbiol Spectr. 2019. PMID: 31441398 Free PMC article. Review. - Stochastic and Differential Activation of σB and PrfA in Listeria monocytogenes at the Single Cell Level under Different Environmental Stress Conditions.
Guldimann C, Guariglia-Oropeza V, Harrand S, Kent D, Boor KJ, Wiedmann M. Guldimann C, et al. Front Microbiol. 2017 Mar 14;8:348. doi: 10.3389/fmicb.2017.00348. eCollection 2017. Front Microbiol. 2017. PMID: 28352251 Free PMC article. - Role of FruR transcriptional regulator in virulence of Listeria monocytogenes and identification of its regulon.
Abdelhamed H, Ramachandran R, Narayanan L, Islam S, Ozan O, Freitag N, Lawrence ML. Abdelhamed H, et al. PLoS One. 2022 Sep 2;17(9):e0274005. doi: 10.1371/journal.pone.0274005. eCollection 2022. PLoS One. 2022. PMID: 36054213 Free PMC article. - Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes.
Mertins S, Joseph B, Goetz M, Ecke R, Seidel G, Sprehe M, Hillen W, Goebel W, Müller-Altrock S. Mertins S, et al. J Bacteriol. 2007 Jan;189(2):473-90. doi: 10.1128/JB.00972-06. Epub 2006 Nov 3. J Bacteriol. 2007. PMID: 17085572 Free PMC article. - Functional impact of mutational activation on the Listeria monocytogenes central virulence regulator PrfA.
Miner MD, Port GC, Freitag NE. Miner MD, et al. Microbiology (Reading). 2008 Nov;154(Pt 11):3579-3589. doi: 10.1099/mic.0.2008/021063-0. Microbiology (Reading). 2008. PMID: 18957610 Free PMC article.
References
- Böckmann, R., C. Dickneite, B. Middendorf, W. Goebel, and Z. Sokolovic. 1996. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol. Microbiol. 22:643-653. - PubMed
- Bubert, A., Z. Sokolovic, S. K. Chun, L. Papatheodorou, A. Simm, and W. Goebel. 1999. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol. Gen. Genet. 261:323-336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources