Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis - PubMed (original) (raw)
Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis
Narito Asanuma et al. Appl Environ Microbiol. 2004 Sep.
Abstract
A ccpA gene that encodes global catabolite control protein A (CcpA) in Streptococcus bovis was identified and characterized, and the involvement of CcpA in transcriptional control of a gene (ldh) encoding lactate dehydrogenase (LDH) and a gene (pfl) encoding pyruvate formate-lyase (PFL) was examined. The ccpA gene was shown to be transcribed as a monocistronic operon. A catabolite-responsive element (cre) was found in the promoter region of ccpA, suggesting that ccpA transcription in S. bovis is autogenously regulated. CcpA required HPr that was phosphorylated at the serine residue at position 46 (HPr-[Ser-P]) for binding to the cre site, but glucose 6-phosphate, fructose 1,6-bisphosphate, and NADP had no effect on binding. Diauxic growth was observed when S. bovis was grown in a medium containing glucose and lactose, but it disappeared when ccpA was disrupted, which indicates that CcpA is involved in catabolite repression in S. bovis. The level of ccpA mRNA was higher when cells were grown on glucose than when they were grown on lactose, which was in line with the level of ldh mRNA. When cells were grown on glucose, the ldh mRNA level was lower but the pfl mRNA level was higher in a ccpA-disrupted mutant than in the parent strain, which suggests that ldh transcription is enhanced and pfl transcription is suppressed by CcpA. The ccpA-disrupted mutant produced less lactate and more formate than the parent, probably because the mutant had reduced LDH activity and elevated PFL activity. In the upper region of both ldh and pfl, a cre-like sequence was found, suggesting that the complex consisting of CcpA and HPr-[Ser-P] binds to the possible cre sites. Thus, CcpA appears to be involved in the global regulation of sugar utilization in S. bovis.
Figures
FIG. 1.
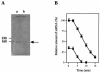
(A) Schematic representation of the ccpA and pepQ genes in S. bovis. A putative transcriptional start site and a termination site for ccpA are indicated by open and solid circles, respectively. A possible cre sequence is shown along with the cre consensus sequence. (B) Primer extension analysis of S. bovis ccpA. Sequence ladders were electrophoresed with the same primer, and the transcriptional start site is indicated by an arrow.
FIG. 2.
(A) Level of ccpA mRNA in S. bovis grown on glucose (lane a) or lactose (lane b), as estimated by Northern blotting. The arrow indicates the position of 1.1-kb ccpA mRNA. The ratio of mRNA levels (level of ccpA mRNA in S. bovis grown on glucose/level of ccpA mRNA in S. bovis grown on lactose) was 3/1. (B) Decay of ccpA mRNA in S. bovis cells grown on glucose (▪) or lactose (•).
FIG. 3.
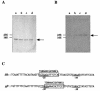
(A) Gel shift assay showing the binding of the complex consisting of CcpA and HPr-[Ser-P] to the cre site. A _cre_-containing fragment was incubated in the presence (plus sign) or in the absence (minus sign) of CcpA, HPr, and/or HPr-[Ser-P]. Bovine serum albumin (BSA) was used as a nonspecific protein. (B) SDS-PAGE of the complex consisting of CcpA and HPr-[Ser-P] after the gel shift assay. Lanes a and b contained CcpA and HPr-[Ser-P], respectively. Lane c contained the complex consisting of CcpA, HPr-[Ser-P], and the _cre_-containing fragment.
FIG. 4.
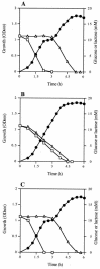
Growth (•) and glucose (□) and lactose (▵) concentrations in the medium when S. bovis 12U1 (A), 12U1-_ccpA_− (B), and 12U1-ccpA+ (C) were grown in a medium containing both glucose and lactose. OD600, optical density at 600 nm.
FIG. 5.
(A) ldh mRNA level in S. bovis 12U1 grown on glucose (lane a) or lactose (lane b) and ldh mRNA level in S. bovis 12U1-_ccpA_− grown on glucose (lane c) or lactose (lane d), as estimated by Northern blotting. The arrow indicates the position of 1.0-kb ldh mRNA. (B) pfl mRNA level in 12U1 grown on glucose (lane a) or lactose (lane b) and pfl mRNA level in 12U1-_ccpA_− grown on glucose (lane c) or lactose (lane d). The arrow indicates the position of 2.3-kb pfl mRNA. (C) Alignment of the ldh (accession number U60997) and pfl (AB014686) promoter regions of S. bovis. The −35 and −10 sequences are underlined. Possible cre sites are enclosed in boxes, and the consensus cre sequence is indicated.
Similar articles
- Structure and transcriptional regulation of the gene encoding pyruvate formate-lyase of a ruminal bacterium, Streptococcus bovis.
Asanuma N, Iwamoto M, Hino T. Asanuma N, et al. Microbiology (Reading). 1999 Jan;145 ( Pt 1):151-157. doi: 10.1099/13500872-145-1-151. Microbiology (Reading). 1999. PMID: 10206694 - Presence of NAD+-specific glyceraldehyde-3-phosphate dehydrogenase and CcpA-dependent transcription of its gene in the ruminal bacterium Streptococcus bovis.
Asanuma N, Hino T. Asanuma N, et al. FEMS Microbiol Lett. 2006 Apr;257(1):17-23. doi: 10.1111/j.1574-6968.2006.00111.x. FEMS Microbiol Lett. 2006. PMID: 16553827 - Carbon catabolite repression by the catabolite control protein CcpA in Staphylococcus xylosus.
Jankovic I, Brückner R. Jankovic I, et al. J Mol Microbiol Biotechnol. 2002 May;4(3):309-14. J Mol Microbiol Biotechnol. 2002. PMID: 11931563 Review. - Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria?
Hueck CJ, Hillen W. Hueck CJ, et al. Mol Microbiol. 1995 Feb;15(3):395-401. doi: 10.1111/j.1365-2958.1995.tb02252.x. Mol Microbiol. 1995. PMID: 7540244 Review.
Cited by
- How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria.
Deutscher J, Francke C, Postma PW. Deutscher J, et al. Microbiol Mol Biol Rev. 2006 Dec;70(4):939-1031. doi: 10.1128/MMBR.00024-06. Microbiol Mol Biol Rev. 2006. PMID: 17158705 Free PMC article. Review. - Properties and role of glyceraldehyde-3-phosphate dehydrogenase in the control of fermentation pattern and growth in a ruminal bacterium, Streptococcus bovis.
Asanuma N, Yoshizawa K, Hino T. Asanuma N, et al. Curr Microbiol. 2009 Apr;58(4):283-7. doi: 10.1007/s00284-008-9326-2. Epub 2008 Nov 25. Curr Microbiol. 2009. PMID: 19034572 - Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae.
Iyer R, Baliga NS, Camilli A. Iyer R, et al. J Bacteriol. 2005 Dec;187(24):8340-9. doi: 10.1128/JB.187.24.8340-8349.2005. J Bacteriol. 2005. PMID: 16321938 Free PMC article. - Proteome analysis of Streptococcus thermophilus grown in milk reveals pyruvate formate-lyase as the major upregulated protein.
Derzelle S, Bolotin A, Mistou MY, Rul F. Derzelle S, et al. Appl Environ Microbiol. 2005 Dec;71(12):8597-605. doi: 10.1128/AEM.71.12.8597-8605.2005. Appl Environ Microbiol. 2005. PMID: 16332852 Free PMC article. - Transcriptome Analysis Reveals Catabolite Control Protein A Regulatory Mechanisms Underlying Glucose-Excess or -Limited Conditions in a Ruminal Bacterium, Streptococcus bovis.
Jin Y, Fan Y, Sun H, Zhang Y, Wang H. Jin Y, et al. Front Microbiol. 2021 Nov 18;12:767769. doi: 10.3389/fmicb.2021.767769. eCollection 2021. Front Microbiol. 2021. PMID: 34867900 Free PMC article.
References
- Asanuma, N., and T. Hino. 1997. Tolerance to low pH and lactate production in rumen bacteria. Anim. Sci. Technol. 68:367-376.
- Asanuma, N., and T. Hino. 2002. Fructose bisphosphate aldolase activity and glycolytic intermediate concentrations in relation to lactate production in Streptococcus bovis. Anaerobe 8:1-8.
- Asanuma, N., and T. Hino. 2003. Molecular characterization of HPr and related enzymes, and regulation of HPr phosphorylation in the ruminal bacterium Streptococcus bovis. Arch. Microbiol. 179:205-213. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources