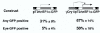Enhancer trapping in zebrafish using the Sleeping Beauty transposon - PubMed (original) (raw)
Enhancer trapping in zebrafish using the Sleeping Beauty transposon
Darius Balciunas et al. BMC Genomics. 2004.
Abstract
Background: Among functional elements of a metazoan gene, enhancers are particularly difficult to find and annotate. Pioneering experiments in Drosophila have demonstrated the value of enhancer "trapping" using an invertebrate to address this functional genomics problem.
Results: We modulated a Sleeping Beauty transposon-based transgenesis cassette to establish an enhancer trapping technique for use in a vertebrate model system, zebrafish Danio rerio. We established 9 lines of zebrafish with distinct tissue- or organ-specific GFP expression patterns from 90 founders that produced GFP-expressing progeny. We have molecularly characterized these lines and show that in each line, a specific GFP expression pattern is due to a single transposition event. Many of the insertions are into introns of zebrafish genes predicted in the current genome assembly. We have identified both previously characterized as well as novel expression patterns from this screen. For example, the ET7 line harbors a transposon insertion near the mkp3 locus and expresses GFP in the midbrain-hindbrain boundary, forebrain and the ventricle, matching a subset of the known FGF8-dependent mkp3 expression domain. The ET2 line, in contrast, expresses GFP specifically in caudal primary motoneurons due to an insertion into the poly(ADP-ribose) glycohydrolase (PARG) locus. This surprising expression pattern was confirmed using in situ hybridization techniques for the endogenous PARG mRNA, indicating the enhancer trap has replicated this unexpected and highly localized PARG expression with good fidelity. Finally, we show that it is possible to excise a Sleeping Beauty transposon from a genomic location in the zebrafish germline.
Conclusions: This genomics tool offers the opportunity for large-scale biological approaches combining both expression and genomic-level sequence analysis using as a template an entire vertebrate genome.
Figures
Figure 1
Artificial enhancer trapping with a Sleeping Beauty transposon. Comparison of GFP expression in embryos injected with pT2/
S1
EF1α or γCry1/pT2/
S1
EF1α. Plasmids are diagramed as cartoons on the top of the picture. The SB transposon's inverted repeats are shown as boxes with open triangles, and the GFP open reading frame is depicted as a grey arrow. The gamma-Crystallin promoter/enhancer is shown as a hatched box. DNA-injected embryos which survived to 3 dpf were counted and scored for GFP fluorescence anywhere in the embryo (any GFP) and for fluorescence in the eye (eye GFP), even if there was additional fluorescence elsewhere. The average percentage of embryos positive for particular GFP fluorescence in three independent experiments is shown ± standard deviation.
Figure 2
EF1α promoter truncations and endogenous enhancer trap screening. A diagram of the
S1
EF1α promoter [32, 41]. Restriction enzyme sites are shown on top as single letters. S is _Sph_I, N is _Nhe_I, B is _Bst_1107I and R is _Eco_RI. G/C, G and C rich box. Sp1, Sp1-like site. TATA, TATA box. Numbering below is relative to the first T of the TATA box. The table below the diagram shows the results of the pilot and scale-up (*) screens. Transgenesis and expression rates are shown, non-expressing transposon insertions were not scored. Transgenesis and expression rate from scale-up screen (#) is an underestimate since many founders were screened by incross and crosses from doubly transgenic founders were scored as a single transmission event (see text).
Figure 3
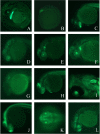
Enhancer trap lines exhibit a variety of unique GFP expression patterns. (A). Lateral view of GFP expression in Enhancer Trap line 1 (ET1) at 38 hours post fertilization (hpf). (B) ET3 at 5–6 somite stage. (C) ET3 at 36 hpf. (D) ET4 at 26 hpf. (E) ET5 at 30 hpf. (F) ET5 at 48 hpf. (G) ET6 at 26 hpf. (H) ET7 at 32 hpf. (I) Ventral view of ET7 at 5 dpf. (J) Lateral view of ET8 at 26 hpf. (K) Dorsal view of ET9 at 28 hpf. (L) Lateral view of ET9 at 30 hpf. In all panels, anterior is to the left. See text for details.
Figure 4
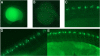
The ET2 transgenic fish line expresses GFP in caudal primary motoneurons. GFP expression in ET2 was visualized in motoneurons using a bandpass GFP filter set at various stages of embryonic development. In all panels anterior is to the left. (A) The onset of GFP expression in ET2 line at 16 somite stage. (B) 26 somite stage. (C) 24 hpf. (D, E) 36 hpf. Axonal trajectories are visible at 24 and 36 hpf.
Figure 5
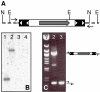
Identification of the transposition event in the ET2 line. (A) The pT2/
S2
EF1α transposon insertion into zebrafish genome is shown; restriction enzyme sites and primers used for molecular analysis are indicated. Transposon IR/DR's are shown as solid boxes with open triangles, and the GFP open reading frame is shown as a grey arrow. Genomic DNA is shown as a dotted line. N is _Nsi_I, E is _Eco_RV. (B) Southern blot on ET2 line outcross embryos. DNA from GFP positive (lanes 1 and 2) and GFP negative (lanes 3 and 4) embryos was digested with _Nsi_I (lanes 1 and 3) or _Eco_RV (lanes 2 and 4) and probed with a GFP-specific probe. (C) Linkage of the transposon insertion event to GFP expression. Primers flanking the transposon insertion event (arrows) were used to conduct PCR on DNA from GFP positive (lane 2) and GFP negative (lane 3) embryos from an ET2 outcross different from the one used in (B). Lane 1, λ _Eco_47III Marker (Fermentas Inc).
Figure 6
GFP expression in ET2 line embryos is indistinguishable from endogenous PARG gene expression. 23 hpf embryos collected from a heterozygous outcross were photographed for GFP fluorescence and sibling embryos were fixed for in situ hybridization. (A) In situ hybridization with PARG antisense probe. (B) In situ with GFP antisense probe. (C) Visualization of GFP expression in living embryos using a bandpass GFP filter set. (D) The same embryo as in (C) photographed using a bandpass GFP filter set with a low level of bright field illumination to visualize GFP expression in relative position to the somites.
Figure 7
GFP expression in ET7 line matches mkp3 mRNA expression. (A) GFP fluorescence photograph of an ET7 embryo at 23 hpf. (B) In situ hybridization on 23 hpf wild type embryo using mkp3 antisense RNA probe.
Similar articles
- Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo.
Parinov S, Kondrichin I, Korzh V, Emelyanov A. Parinov S, et al. Dev Dyn. 2004 Oct;231(2):449-59. doi: 10.1002/dvdy.20157. Dev Dyn. 2004. PMID: 15366023 - [Enhancer trapping nearby rps26 gene in zebrafish mediated by the Tol2 transposon and it's annotation].
Sang Y, Shen D, Chen W, Chan S, Gu H, Gao B, Song C. Sang Y, et al. Sheng Wu Gong Cheng Xue Bao. 2018 Mar 25;34(3):449-458. doi: 10.13345/j.cjb.170306. Sheng Wu Gong Cheng Xue Bao. 2018. PMID: 29577695 Chinese. - A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish.
Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. Kawakami K, et al. Dev Cell. 2004 Jul;7(1):133-44. doi: 10.1016/j.devcel.2004.06.005. Dev Cell. 2004. PMID: 15239961 - Transposon tools and methods in zebrafish.
Kawakami K. Kawakami K. Dev Dyn. 2005 Oct;234(2):244-54. doi: 10.1002/dvdy.20516. Dev Dyn. 2005. PMID: 16110506 Review.
Cited by
- New tools for the identification of developmentally regulated enhancer regions in embryonic and adult zebrafish.
Levesque MP, Krauss J, Koehler C, Boden C, Harris MP. Levesque MP, et al. Zebrafish. 2013 Mar;10(1):21-9. doi: 10.1089/zeb.2012.0775. Epub 2013 Mar 5. Zebrafish. 2013. PMID: 23461416 Free PMC article. - Labelling and targeted ablation of specific bipolar cell types in the zebrafish retina.
Zhao XF, Ellingsen S, Fjose A. Zhao XF, et al. BMC Neurosci. 2009 Aug 27;10:107. doi: 10.1186/1471-2202-10-107. BMC Neurosci. 2009. PMID: 19712466 Free PMC article. - Hierarchical control of locomotion by distinct types of spinal V2a interneurons in zebrafish.
Menelaou E, McLean DL. Menelaou E, et al. Nat Commun. 2019 Sep 13;10(1):4197. doi: 10.1038/s41467-019-12240-3. Nat Commun. 2019. PMID: 31519892 Free PMC article. - Efficient disruption of Zebrafish genes using a Gal4-containing gene trap.
Balciuniene J, Nagelberg D, Walsh KT, Camerota D, Georlette D, Biemar F, Bellipanni G, Balciunas D. Balciuniene J, et al. BMC Genomics. 2013 Sep 14;14:619. doi: 10.1186/1471-2164-14-619. BMC Genomics. 2013. PMID: 24034702 Free PMC article. - The zebrafish anatomy and stage ontologies: representing the anatomy and development of Danio rerio.
Van Slyke CE, Bradford YM, Westerfield M, Haendel MA. Van Slyke CE, et al. J Biomed Semantics. 2014 Feb 25;5(1):12. doi: 10.1186/2041-1480-5-12. J Biomed Semantics. 2014. PMID: 24568621 Free PMC article.
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, Szustakowki J, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, International Human Genome Sequencing C Initial sequencing and analysis of the human genome.[see comment][erratum appears in Nature 2001 Aug 2;412(6846):565 Note: Szustakowki J [corrected to Szustakowski J]] Nature. 2001;409:860–921. doi: 10.1038/35057062. - DOI - PubMed
- Venter JC, Adams MD, Myers EWLiPW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome.[see comment][erratum appears in Science 2001 Jun 5;292(5523):1838] Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. - DOI - PubMed
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES, Mouse Genome Sequencing C Initial sequencing and comparative analysis of the mouse genome.[see comment] Nature. 2002;420:520–562. doi: 10.1038/nature01262. - DOI - PubMed
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera , Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Cooney AJ, D'Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Alba M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hubner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Payseur BA, Bourque G, Lopez-Otin C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F, Rat Genome Sequencing Project C Genome sequence of the Brown Norway rat yields insights into mammalian evolution.[see comment] Nature. 2004;428:493–521. doi: 10.1038/nature02426. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous