Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast - PubMed (original) (raw)
Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast
Pil Jung Kang et al. Mol Biol Cell. 2004 Nov.
Abstract
In the budding yeast Saccharomyces cerevisiae, selection of the bud site determines the axis of polarized cell growth and eventual oriented cell division. Bud sites are selected in specific patterns depending on cell type. These patterns appear to depend on distinct types of marker proteins in the cell cortex; in particular, the bipolar budding of diploid cells depends on persistent landmarks at the birth-scar-distal and -proximal poles that involve the proteins Bud8p and Bud9p, respectively. Rax1p and Rax2p also appear to function specifically in bipolar budding, and we report here a further characterization of these proteins and of their interactions with Bud8p and Bud9p. Rax1p and Rax2p both appear to be integral membrane proteins. Although commonly used programs predict different topologies for Rax2p, glycosylation studies indicate that it has a type I orientation, with its long N-terminal domain in the extracytoplasmic space. Analysis of rax1 and rax2 mutant budding patterns indicates that both proteins are involved in selecting bud sites at both the distal and proximal poles of daughter cells as well as near previously used division sites on mother cells. Consistent with this, GFP-tagged Rax1p and Rax2p were both observed at the distal pole as well as at the division site on both mother and daughter cells; localization to the division sites was persistent through multiple cell cycles. Localization of Rax1p and Rax2p was interdependent, and biochemical studies showed that these proteins could be copurified from yeast. Bud8p and Bud9p could also be copurified with Rax1p, and localization studies provided further evidence of interactions. Localization of Rax1p and Rax2p to the bud tip and distal pole depended on Bud8p, and normal localization of Bud8p was partially dependent on Rax1p and Rax2p. Although localization of Rax1p and Rax2p to the division site did not appear to depend on Bud9p, normal localization of Bud9p appeared largely or entirely dependent on Rax1p and Rax2p. Taken together, the results indicate that Rax1p and Rax2p interact closely with each other and with Bud8p and Bud9p in the establishment and/or maintenance of the cortical landmarks for bipolar budding.
Figures
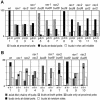
Figure 1.
Budding patterns of diploid strains homozygous for various mutations. At least 300 cells were scored for each type of count on each strain, and percentages are indicated. Strains used were wild-type (YEF473), rax1 (HPY496), rax2 (HPY592), rax1 rax2 (HPY634), bud8 (YHH415), rax1 bud8 (HPY643), rax2 bud8 (HPY641), bud9 (YHH615), rax1 bud9 (HPY692), rax2 bud9 (HPY693), rax1 axl2 (HPY841), and rax2 axl2 (HPY842). (A) The positions of the first buds on daughter cells were scored as being at the pole proximal to the birth scar (p), at the pole distal to the birth scar (d), or in the middle region (m) of the cell. (B) The budding patterns of the strains were determined by counting cells that had >3 bud scars. (a) One or more chains of bud scars similar to those of the axial pattern; (b) bud scars concentrated at one or both poles of the cell; (r) one or more bud scars at seemingly random sites. Within category b, the gray, black, and hatched boxes indicate cells with bud scars at both poles, exclusively at the distal pole, or exclusively at the proximal pole, respectively.
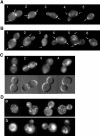
Figure 2.
Interdependent localization of Rax1p and Rax2p to both the division site and the distal pole. (A-C) Wild-type diploid cells expressing Rax2p-GFP (A; strain HPY814) or GFP-Rax1p (B and C; strain HPY614) were examined for the localization of GFP fluorescence at various stages in the cell cycle. Cells are numbered for reference in the text; arrows indicate signal at previous division sites. In C, the companion DIC images show the presence (cells 1 and 2) or absence (cells 3 and 4) of a fully formed septum. (D) Diploid cells expressing Rax2p-GFP in a _rax1_Δ background (a; strain HPY836) or GFP-Rax1p in a _rax2_Δ background (b; strain HPY647) were examined for the localization of GFP fluorescence.
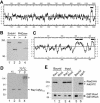
Figure 3.
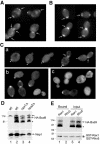
Biochemical properties and interaction of Rax1p and Rax2p. (A) Hydropathy plot of Rax2p generated by the method of Kyte and Doolittle (1982) with a window size of 11. The region N-terminal to the predicted transmembrane domain (black bar) contains 21% S + T and 52 potential N-linked glycosylation sites (N-X-S/T, where X is any amino acid other than P; Tanner and Lehle, 1987). (B) _N_-linked glycosylation of Rax2p. Total cell lysates from rax2_Δ strain AM476 containing RAX2-HA 3 plasmid pHP1183 were either mock-digested (-) or digested with Endo H or PNGase (+) before SDS-PAGE and immunoblotting with anti-HA antibody 3F10. (C) Hydropathy plot of Rax1p generated as described in A. Black bars, possible transmembrane domains. (D) Mobility of Rax1p-Myc13 in SDS-PAGE and its aggregation upon heating in SDS solution. Immunoblotting was performed using anti-Myc antibody 9E10 (see Materials and Methods). Lanes 1 and 2, proteins from total cell lysates of wild-type strain HPY16 (lane 1) or RAX1-MYC 13 strain HPY843 (lane 2) were mixed with SDS-PAGE sample buffer and loaded onto a 6% gel after a 5-min incubation at room temperature (r.t.). Lanes 3 and 4, total proteins from HPY843 were mixed with SDS-PAGE sample buffer and heated at 70°C for 1 (lane 3) or 5 (lane 4) min before loading onto the gel. The asterisk indicates the aggregated Rax1p-Myc13. (E) Copurification of Rax1p and Rax2p from yeast. Cells of strain HPY644 (RAX2-HA 3) carrying GAL1p-GST-HIS 6 -RAX1 plasmid pHP1156 (lanes 1 and 3) or GAL1p-GST-HIS 6 -RHO5 plasmid pHP1158 (lanes 2 and 4) were grown in SC-Ura medium with 2% sucrose as carbon source and then induced for 4 h by adding galactose to 2%. The membrane fractions were isolated, solubilized with 1% Triton X-100, and subjected to pull-down of the GST-tagged protein (see Materials and Methods). Samples of the proteins eluted from the glutathione Sepharose (lanes 1 and 2) and of the input material (lanes 3 and 4) were analyzed by SDS-PAGE and immunoblotting with anti-HA antibody HA11 (top panel) and anti-GST antibody (bottom panel; see Materials and Methods). As a control (lanes 5 and 6), the membrane fraction prepared from cells of strain HPY798 (rax1_Δ) containing plasmids pHP1169 (AXL2-PC) and pHP1156 was analyzed similarly using antibodies against protein C (top panel) and GST (bottom panel).
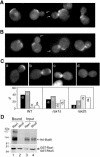
Figure 4.
Interaction between Rax1p/Rax2p and Bud8p. (A and B) Localization of Rax1p and Rax2p to the bud tip depends on Bud8p. Cells of _GFP-RAX1 bud8_Δ strain HPY690 (A) and _RAX2-GFP bud8_Δ strain HPY691 (B) were examined for the localization of GFP fluorescence. Arrows indicate bud tips lacking GFP-Rax1p or Rax2p-GFP; localization to mother-bud necks and previous division sites appears normal. (C) Efficient localization of Bud8p to the bud tip depends on Rax1p and Rax2p. The _bud8_Δ strain YHH415 (WT), _rax1_Δ _bud8_Δ strain HPY643 (_rax1_Δ), and _rax2_Δ _bud8_Δ strain HPY641 (_rax2_Δ) were transformed with GFP-BUD8 plasmid YEpGFP-BUD8F, and ∼100 large-budded cells were examined for the localization of GFP fluorescence. Four patterns of GFP-Bud8p localization were observed, as shown in the representative images: (a) fluorescence exclusively at or near the bud tip; (b) fluorescence at both the bud tip and neck; (c) fluorescence exclusively at the neck (often specifically on the daughter side); and (d) no localized fluorescence. The percentage of each pattern of each strain is shown in the histograms. Note that the high perentage of wild-type cells with GFP-Bud8p at the neck as well as the distal pole probably reflects the overexpression of GFP-Bud8p in these cells (Harkins et al., 2001; Schenkman et al., 2002). (D) Copurification of Rax1p and Bud8p from yeast. Cells of _bud8_Δ strain HPY568 carrying HA 3 -BUD8 plasmid YEpHA-BUD8F and either GAL1p-GST-HIS 6 -RAX1 plasmid pHP1156 (lanes 1 and 3) or GAL1p-GST-HIS 6 -RHO5 plasmid pHP1158 (lanes 2 and 4) were grown and analyzed as described for Figure 3E.
Figure 5.
Interaction between Rax1p/Rax2p and Bud9p. (A and B) Localization of Rax1p and Rax2p does not appear to depend on Bud9p. Cells of _GFP-RAX1 bud9_Δ strain HPY704 (A) and _RAX2-GFP bud9_Δ strain HPY705 (B) were examined for the localization of GFP fluorescence. Arrows indicate seemingly normal localization of GFP-Rax1p and Rax2p-GFP to mother-bud necks and previous division sites. (C) Localization of Bud9p depends on Rax1p and Rax2p. Wild-type strain YEF473 (a), _rax1_Δ strain HPY496 (b), and _rax2_Δ strain HPY592 (c) were transformed with GFP 3 -BUD9 plasmid pHP1202, and cells were examined for the localization of GFP fluorescence. In panel a, asterisks indicate the proximal poles as judged by Calcofluor staining, and arrows indicate GFP3-Bud9p localized normally to the daughter side of the neck. (D) Bud9p levels are approximately normal in the absence of Rax1p or Rax2p. Wild-type strain YEF473 (lane 2), _rax1_Δ strain HPY496 (lane 3), and _rax2_Δ strain HPY592 (lane 4) were transformed with HA 3 -BUD9 plasmid pHP1319, and total cell lysates from the same OD units of cells were analyzed by immunoblotting using anti-HA antibody HA11 (top panel) or anti-Nap1p antibodies as a loading control (bottom panel). As a control, total cell lysate was prepared from YEF473 without plasmid and subjected to the same procedure (lane 1). The asterisk indicates a presumably degraded or undermodified Bud9p; the double asterisk indicates a protein cross-reacting nonspecifically with the anti-HA antibody. (E) Copurification of Rax1p and Bud9p from yeast. Cells of _bud9_Δ strain HPY570 carrying HA 3 -BUD9 plasmid pHP1319 and either GAL1p-GST-HIS 6 -RAX1 plasmid pHP1156 (lanes 1 and 3) or GAL1p-GST-HIS 6 -RHO5 plasmid pHP1158 (lanes 2 and 4) were grown and analyzed as described for Figure 3E. The asterisk indicates a presumably degraded or undermodified Bud9p.
References
- Adames, N., Blundell, K., Ashby, M.N., and Boone, C. (1995). Role of yeast insulin-degrading enzyme homologs in propheromone processing and bud site selection. Science 270, 464-467. -PubMed
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., and Struhl, K. (1999). Current Protocols in Molecular Biology, New York: John Wiley & Sons.
- Chant, J., and Herskowitz, I. (1991). Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65, 1203-1212. -PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM56997/GM/NIGMS NIH HHS/United States
- R37 GM031006/GM/NIGMS NIH HHS/United States
- GM31006/GM/NIGMS NIH HHS/United States
- R01 GM056997/GM/NIGMS NIH HHS/United States
- R01 GM031006/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases