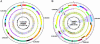Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis - PubMed (original) (raw)
. 2004 Sep 21;101(38):13826-31.
doi: 10.1073/pnas.0404012101. Epub 2004 Sep 9.
E Carniel, F W Larimer, J Lamerdin, P O Stoutland, W M Regala, A M Georgescu, L M Vergez, M L Land, V L Motin, R R Brubaker, J Fowler, J Hinnebusch, M Marceau, C Medigue, M Simonet, V Chenal-Francisque, B Souza, D Dacheux, J M Elliott, A Derbise, L J Hauser, E Garcia
Affiliations
- PMID: 15358858
- PMCID: PMC518763
- DOI: 10.1073/pnas.0404012101
Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis
P S G Chain et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Yersinia pestis, the causative agent of plague, is a highly uniform clone that diverged recently from the enteric pathogen Yersinia pseudotuberculosis. Despite their close genetic relationship, they differ radically in their pathogenicity and transmission. Here, we report the complete genomic sequence of Y. pseudotuberculosis IP32953 and its use for detailed genome comparisons with available Y. pestis sequences. Analyses of identified differences across a panel of Yersinia isolates from around the world reveal 32 Y. pestis chromosomal genes that, together with the two Y. pestis-specific plasmids, to our knowledge, represent the only new genetic material in Y. pestis acquired since the the divergence from Y. pseudotuberculosis. In contrast, 149 other pseudogenes (doubling the previous estimate) and 317 genes absent from Y. pestis were detected, indicating that as many as 13% of Y. pseudotuberculosis genes no longer function in Y. pestis. Extensive insertion sequence-mediated genome rearrangements and reductive evolution through massive gene loss, resulting in elimination and modification of preexisting gene expression pathways, appear to be more important than acquisition of genes in the evolution of Y. pestis. These results provide a sobering example of how a highly virulent epidemic clone can suddenly emerge from a less virulent, closely related progenitor.
Figures
Fig. 1.
Circular genome map of IP32953 and comparison with Y. pestis CO92. (A) Genome of IP32953. (B) Genome of CO92. (A and B) Circle 1 (from center outward), G+C content; circles 2 and 3, all genes coded by function (forward and reverse strand); circle 4, GC skew (G–C/G+C); circles 5 and 6, genome divided into locally colinear blocks (when IP32953 and CO92 are compared with one another); each block is distinguished by a unique color (black segments within colored blocks represent regions specific to that genome in the comparison), and the orientation of each block is indicated by strand; [circle 5, –ve strand; circle 6, +ve strand); circle 7, locations of IS elements (IS_100_ is blue, IS_285_ is red, IS_1661_ is green, and IS_1541_ is magenta)]. In A, the gray highlighted region near the 12 o'clock position indicates the proposed IP32953 inversion (see text), whereas the remainder of the genome denotes the stable “ancestral” arrangement that has prevailed through the present. B illustrates the complexity of the molecular events that gave rise to the inversions or translocations in the Y. pestis genome first proposed (16) solely on the basis of the dramatic shifts in G/C skew (gray highlights serotypes I, II, and III), but now extended through whole-genome comparison. For example, gray highlight II is composed of three distinct blocks, two that are derived from distinct places within the same replichore (origin to terminus half), whereas the third block originated from the other replichore (light blue block).
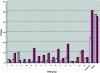
Fig. 2.
Functional classification of genes missing or inactivated in Y. pestis. Distribution of _Y. pestis_-specific lost functions by gene region deletion (light blue) or by gene inactivation (i.e., pseudogene, dark purple) in COG functional groups: C, energy production; D, cell division and/or chromosome partitioning; E, amino acid metabolism; F, nucleotide metabolism; G, carbohydrate metabolism; H, coenzyme metabolism; I, lipid metabolism; J, translation; K, transcription; L, DNA replication and/or repair; M, cell envelope biogenesis; N, cell motility, secretion; O, posttranslational modification; P, inorganic ion metabolism; R, general function prediction only; S, function unknown; T, signal transduction; conserved, conserved hypothetical genes with no significant COG hits; and unique, hypothetical genes with no significant COG hit.
Similar articles
- Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis.
Hinchliffe SJ, Isherwood KE, Stabler RA, Prentice MB, Rakin A, Nichols RA, Oyston PC, Hinds J, Titball RW, Wren BW. Hinchliffe SJ, et al. Genome Res. 2003 Sep;13(9):2018-29. doi: 10.1101/gr.1507303. Genome Res. 2003. PMID: 12952873 Free PMC article. - Characterization of chromosomal regions conserved in Yersinia pseudotuberculosis and lost by Yersinia pestis.
Pouillot F, Fayolle C, Carniel E. Pouillot F, et al. Infect Immun. 2008 Oct;76(10):4592-9. doi: 10.1128/IAI.00568-08. Epub 2008 Aug 4. Infect Immun. 2008. PMID: 18678673 Free PMC article. - The complete genome sequence of Yersinia pseudotuberculosis IP31758, the causative agent of Far East scarlet-like fever.
Eppinger M, Rosovitz MJ, Fricke WF, Rasko DA, Kokorina G, Fayolle C, Lindler LE, Carniel E, Ravel J. Eppinger M, et al. PLoS Genet. 2007 Aug;3(8):e142. doi: 10.1371/journal.pgen.0030142. Epub 2007 Jul 10. PLoS Genet. 2007. PMID: 17784789 Free PMC article. - Small Insertions and Deletions Drive Genomic Plasticity during Adaptive Evolution of Yersinia pestis.
Wu Y, Hao T, Qian X, Zhang X, Song Y, Yang R, Cui Y. Wu Y, et al. Microbiol Spectr. 2022 Jun 29;10(3):e0224221. doi: 10.1128/spectrum.02242-21. Epub 2022 Apr 19. Microbiol Spectr. 2022. PMID: 35438532 Free PMC article. Review. - Current trends in plague research: from genomics to virulence.
Huang XZ, Nikolich MP, Lindler LE. Huang XZ, et al. Clin Med Res. 2006 Sep;4(3):189-99. doi: 10.3121/cmr.4.3.189. Clin Med Res. 2006. PMID: 16988099 Free PMC article. Review.
Cited by
- Gene Loss Dominates As a Source of Genetic Variation within Clonal Pathogenic Bacterial Species.
Bolotin E, Hershberg R. Bolotin E, et al. Genome Biol Evol. 2015 Jul 10;7(8):2173-87. doi: 10.1093/gbe/evv135. Genome Biol Evol. 2015. PMID: 26163675 Free PMC article. - Evolution and virulence contributions of the autotransporter proteins YapJ and YapK of Yersinia pestis CO92 and their homologs in Y. pseudotuberculosis IP32953.
Lenz JD, Temple BR, Miller VL. Lenz JD, et al. Infect Immun. 2012 Oct;80(10):3693-705. doi: 10.1128/IAI.00529-12. Epub 2012 Jul 16. Infect Immun. 2012. PMID: 22802344 Free PMC article. - An Atypical AAA+ ATPase Assembly Controls Efficient Transposition through DNA Remodeling and Transposase Recruitment.
Arias-Palomo E, Berger JM. Arias-Palomo E, et al. Cell. 2015 Aug 13;162(4):860-71. doi: 10.1016/j.cell.2015.07.037. Cell. 2015. PMID: 26276634 Free PMC article. - A North American Yersinia pestis draft genome sequence: SNPs and phylogenetic analysis.
Touchman JW, Wagner DM, Hao J, Mastrian SD, Shah MK, Vogler AJ, Allender CJ, Clark EA, Benitez DS, Youngkin DJ, Girard JM, Auerbach RK, Beckstrom-Sternberg SM, Keim P. Touchman JW, et al. PLoS One. 2007 Feb 21;2(2):e220. doi: 10.1371/journal.pone.0000220. PLoS One. 2007. PMID: 17311096 Free PMC article. - A Recombinant Attenuated Yersinia pseudotuberculosis Vaccine Delivering a Y. pestis YopENt138-LcrV Fusion Elicits Broad Protection against Plague and Yersiniosis in Mice.
Singh AK, Curtiss R 3rd, Sun W. Singh AK, et al. Infect Immun. 2019 Sep 19;87(10):e00296-19. doi: 10.1128/IAI.00296-19. Print 2019 Oct. Infect Immun. 2019. PMID: 31331960 Free PMC article.
References
- Brenner, D. J., Steigerwalt, A. G., Falcao, D. P., Weaver, R. E. & Fanning, G. R. (1976) Int. J. Syst. Bacteriol. 26, 180–194.
- Moore, R. L. & Brubaker, R. R. (1975) Int. J. Syst. Bacteriol. 25, 336–339.
- Cornelis, G. R. & Van Gijsegem, F. (2000) Annu. Rev. Microbiol. 54, 735–774. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases