Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1 - PubMed (original) (raw)
Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1
Youssef Belkhadir et al. Plant Cell. 2004 Oct.
Abstract
Bacterial pathogens deliver type III effector proteins into the plant cell during infection. On susceptible (r) hosts, type III effectors can contribute to virulence. Some trigger the action of specific disease resistance (R) gene products. The activation of R proteins can occur indirectly via modification of a host target. Thus, at least some type III effectors are recognized at site(s) where they may act as virulence factors. These data indicate that a type III effector's host target might be required for both initiation of R function in resistant plants and pathogen virulence in susceptible plants. In Arabidopsis thaliana, RPM1-interacting protein 4 (RIN4) associates with both the Resistance to Pseudomonas syringae pv maculicola 1 (RPM1) and Resistance to P. syringae 2 (RPS2) disease resistance proteins. RIN4 is posttranslationally modified after delivery of the P. syringae type III effectors AvrRpm1, AvrB, or AvrRpt2 to plant cells. Thus, RIN4 may be a target for virulence functions of these type III effectors. We demonstrate that RIN4 is not the only host target for AvrRpm1 and AvrRpt2 in susceptible plants because its elimination does not diminish their virulence functions. In fact, RIN4 negatively regulates AvrRpt2 virulence function. RIN4 also negatively regulates inappropriate activation of both RPM1 and RPS2. Inappropriate activation of RPS2 is nonspecific disease resistance 1 (NDR1) independent, in contrast with the established requirement for NDR1 during AvrRpt2-dependent RPS2 activation. Thus, RIN4 acts either cooperatively, downstream, or independently of NDR1 to negatively regulate RPS2 in the absence of pathogen. We propose that many P. syringae type III effectors have more than one target in the host cell. We suggest that a limited set of these targets, perhaps only one, are associated with R proteins. Thus, whereas any pathogen virulence factor may have multiple targets, the perturbation of only one is necessary and sufficient for R activation.
Figures
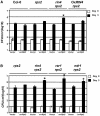
Figure 1.
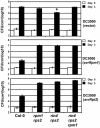
RPM1 Function Is Abrogated in rin4 Null Plants. Growth of the Pto DC3000 strains expressing the indicated type III effector genes, displayed on the right, was measured on wild-type and mutant Arabidopsis lines indicated at the bottom. Four-week-old plants were infiltrated with 105 colony-forming units (cfu)/mL and the number of bacteria per area of leaf plotted on a log10 scale for day 0 (open bars) and day 3 (closed bars) (see Methods). Error bars represent the standard deviation among four samples. This experiment is representative of four independent replicates. The absence of error bars indicates low errors. A one-way analysis of variance (ANOVA) test was applied to each pair of values, and P < 0.01 for rin4 rps2 inoculated with Pto DC3000 (vector) compared with all of the others (asterisk).
Figure 2.
Residual RPM1 Is Sufficient for Constitutive Defense Response in rin4 Null Plants. (A) Total protein extracts were prepared from wild-type Col-0, rpm1 rps2, rin4 rps2, rin4 rps2 rpm1, Ws-0, and rin4 knock-down (rin4K-D) plants. These extracts were subjected to anti-RIN4 (top, WB:RIN4) or anti-PR1 (middle, WB:PR1) protein gel blot analysis. Ponceau staining of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco; bottom) was for confirmation of equal loading in each lane. This experiment is representative of at least three independent replicates. The models summarize the protein gel blot data. Gray shapes represent the plasma membrane. Red shapes represent RPM1 and RPS2 potentially in complex with other cellular proteins, light and dark blue. In rin4 null plants (left), RPM1 and RPS2 are inappropriately activated in the absence of pathogens. In rin4 rps2 plants (right), the residual RPM1 present is activated by the lack of RIN4. The pale blue and red arrows represent RPM1 and RPS2 activation, respectively. The levels of activation are proportional to the thickness of the arrows. (B) Total protein extracts were prepared from wild-type Ws-0 and isogenic RPM1-myc, rin4K-D, and rin4K-D RPM1-myc plants. These extracts were subjected to anti-RIN4 (top, WB:RIN4), anti-PR1 (middle, WB:PR1), and anti-myc (bottom, WB:myc) protein gel blots. Ponceau staining of ribulose-1,5-bisphosphate carboxylase/oxygenase (middle two panels) demonstrates equal loading in each lane for the anti-RIN4 and anti-PR1 antibodies. For the myc protein gel blot, the nonspecific band detected below RPM1-myc was used as an equal loading control. Note that the PR1 immunoblot in (A) is slightly overexposed relative to that in (B). This experiment is indicative of three independent replicates. The models (symbols as in [A]) show that RPM1 and RPS2 are inappropriately active when levels of RIN4 are lowered in rin4K-D. When more RPM1 is expressed (right, note bigger red RPM1 in model), it expresses a higher amplitude of inappropriate activation.
Figure 3.
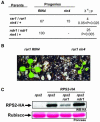
RAR1, but Not NDR1, Delays the Lethality in rin4 Null Plants. (A) F2 plants of the genotypes shown at left were allowed to self-pollinate. The segregation of RIN4 in these progenies was scored on 100 F3 plants by RIN4 protein gel blot analysis. Segregation data were evaluated with χ2 analysis. (B) Representative progenies from selfed RIN4/rin4 rar1/rar1 F2 plants. Note that rar1 rin4 are smaller and develop spontaneous lesions compared with rar1 RIN4 plants. (C) Total protein extracts were prepared from the genotypes listed at the top. These extracts were subjected to anti-HA protein gel blot analysis (top). The Ponceau stain of the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco; bottom) shows that the differences observed in rar1 rps2 (RPS2-HA) plants are not because of loading errors.
Figure 4.
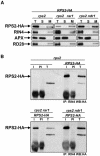
Microsomal RPS2 Localization and Interaction with RIN4 Do Not Require NDR1 or RAR1. (A) Total protein extracts (T) from genotypes shown at the top were fractionated into soluble (S) and microsomal (M) extracts (see Methods). The fractionated samples were analyzed by protein gel blots with anti-HA, anti-RIN4, anti-APX (ascorbate peroxidase; control soluble protein), and anti-RD28 (control integral membrane protein) antisera (Boyes et al., 1998). Microsomal fractions are approximately five times concentrated relative to total and soluble fractions. (B) Protein from genotypes shown at top were immunoprecipitated (IP: RIN4) with anti-RIN4 sera (I) or with preimmune sera (PI). Total extracts (T) from rps2 and rps2 (RPS2-HA) as well as immunoprecipitated samples were analyzed by protein gel blots with an anti-HA antibody (WB: HA). The relative amounts of protein from the immune pellet and the total extracts are not equivalent. The pellet is overrepresented by 30-fold. This experiment is representative of two independent replicates.
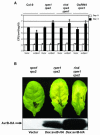
Figure 5.
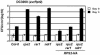
Enhanced RPS2 Function Modulates Its Requirement for RAR1 but Does Not Overcome Its Requirement for NDR1. Growth of Pto DC3000 (avrRpt2) was measured on wild-type and mutant Arabidopsis lines indicated at the bottom. Bacterial growth was measured as described in the legend of Figure 1. Error bars represent the standard deviation among four samples, and this experiment is representative of two independent replicates. The absence of error bars indicates low errors.
Figure 6.
RIN4, RAR1, and NDR1 Modulate AvrRpt2 Virulence Function(s). (A) RIN4 is not required for AvrRpt2 virulence function. Growth of Pma M6CΔE carrying either empty vector or avrRpt2 (indicated at bottom) was measured on the genotypes indicated at top. Bacterial growth was measured as described in the legend of Figure 1. A one-way ANOVA test was applied to each pair of values, and P < 0.01 for rin4 rps2 inoculated with Pma M6CΔE (avrRpt2) compared with all others (asterisks). Error bars represent the standard deviation among four samples, and this experiment is representative of six independent replicates. (B) RAR1 and NDR1 negatively regulate AvrRpt2 virulence function. Inoculations and labels are as in (A). A one-way ANOVA test was applied to each pair of values, and P < 0.01 for rin4 rps2, ndr1 rps2, and rar1 rps2 inoculated with Pma M6CΔE (avrRpt2) compared with all the others (asterisks). Error bars represent the standard deviation among four samples, and this experiment is representative of two independent replicates.
Figure 7.
RIN4 Is Not the Only Virulence Target for AvrRpm1 and AvrB in Arabidopsis. (A) Growth of Pma M6CΔE carrying empty vector or avrRpm1 indicated at bottom was measured on the genotypes indicated at top. Four-week-old plants were infiltrated with 104 cfu/mL and the number of bacteria per area of leaf plotted on a log10 scale for day 0 (open bars) and day 3 (closed bars) (see Methods). Error bars represent the standard deviation among four samples, and this experiment is representative of three independent replicates. The absence of error bars indicates insignificant differences. (B) Agrobacterium tumefaciens carrying empty vector or dexamethasone (DEX) inducible avrB-HA as indicated at bottom were inoculated onto leaves of various genotypes indicated at top, at 1010 cfu/mL. Leaves were sprayed 24 h postinoculation with DEX (20 μM) and photographed 96 h after that. Total protein extracts were prepared 96 h after DEX and subjected to anti-HA protein gel blot analysis.
Similar articles
- The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation.
Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. Kim HS, et al. Proc Natl Acad Sci U S A. 2005 May 3;102(18):6496-501. doi: 10.1073/pnas.0500792102. Epub 2005 Apr 21. Proc Natl Acad Sci U S A. 2005. PMID: 15845764 Free PMC article. - NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis.
Day B, Dahlbeck D, Staskawicz BJ. Day B, et al. Plant Cell. 2006 Oct;18(10):2782-91. doi: 10.1105/tpc.106.044693. Epub 2006 Sep 29. Plant Cell. 2006. PMID: 17012600 Free PMC article. - AvrRpm1 missense mutations weakly activate RPS2-mediated immune response in Arabidopsis thaliana.
Cherkis KA, Temple BR, Chung EH, Sondek J, Dangl JL. Cherkis KA, et al. PLoS One. 2012;7(8):e42633. doi: 10.1371/journal.pone.0042633. Epub 2012 Aug 6. PLoS One. 2012. PMID: 22880057 Free PMC article. - Plant defense: one post, multiple guards?!
Marathe R, Dinesh-Kumar SP. Marathe R, et al. Mol Cell. 2003 Feb;11(2):284-6. doi: 10.1016/s1097-2765(03)00072-8. Mol Cell. 2003. PMID: 12620215 Review. - Role of RIN4 in Regulating PAMP-Triggered Immunity and Effector-Triggered Immunity: Current Status and Future Perspectives.
Ray SK, Macoy DM, Kim WY, Lee SY, Kim MG. Ray SK, et al. Mol Cells. 2019 Jul 31;42(7):503-511. doi: 10.14348/molcells.2019.2433. Mol Cells. 2019. PMID: 31362467 Free PMC article. Review.
Cited by
- Climate change impedes plant immunity mechanisms.
Son S, Park SR. Son S, et al. Front Plant Sci. 2022 Nov 29;13:1032820. doi: 10.3389/fpls.2022.1032820. eCollection 2022. Front Plant Sci. 2022. PMID: 36523631 Free PMC article. Review. - RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean.
Selote D, Kachroo A. Selote D, et al. Plant Physiol. 2010 Jul;153(3):1199-211. doi: 10.1104/pp.110.158147. Epub 2010 May 18. Plant Physiol. 2010. PMID: 20484023 Free PMC article. - The Upregulated Expression of the Citrus RIN4 Gene in HLB Diseased Citrus Aids Candidatus Liberibacter Asiaticus Infection.
Cheng C, Zhong Y, Wang B, Zhang Y, Wu H, Jiang N, Wu B, Lv Y, Jiang B. Cheng C, et al. Int J Mol Sci. 2022 Jun 23;23(13):6971. doi: 10.3390/ijms23136971. Int J Mol Sci. 2022. PMID: 35805971 Free PMC article. - Post-translational modification of host proteins in pathogen-triggered defence signalling in plants.
Stulemeijer IJ, Joosten MH. Stulemeijer IJ, et al. Mol Plant Pathol. 2008 Jul;9(4):545-60. doi: 10.1111/j.1364-3703.2008.00468.x. Mol Plant Pathol. 2008. PMID: 18705867 Free PMC article. Review. - RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector.
Bernoux M, Timmers T, Jauneau A, Brière C, de Wit PJ, Marco Y, Deslandes L. Bernoux M, et al. Plant Cell. 2008 Aug;20(8):2252-64. doi: 10.1105/tpc.108.058685. Epub 2008 Aug 15. Plant Cell. 2008. PMID: 18708476 Free PMC article.
References
- Abramovitch, R., and Martin, G.B. (2004). Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7, 356–364. - PubMed
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. - PubMed
- Austin, M.J., Muskett, P.J., Kahn, K., Feys, B.J., Jones, J.D.G., and Parker, J.E. (2002). Regulatory role of SGT1 in early R-mediated plant defenses. Science 295, 2077–2080. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous