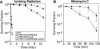Preserving genome integrity: the DdrA protein of Deinococcus radiodurans R1 - PubMed (original) (raw)
Preserving genome integrity: the DdrA protein of Deinococcus radiodurans R1
Dennis R Harris et al. PLoS Biol. 2004 Oct.
Erratum in
- PLoS Biol. 2006 Nov;4(11):e385
Abstract
The bacterium Deinococcus radiodurans can withstand extraordinary levels of ionizing radiation, reflecting an equally extraordinary capacity for DNA repair. The hypothetical gene product DR0423 has been implicated in the recovery of this organism from DNA damage, indicating that this protein is a novel component of the D. radiodurans DNA repair system. DR0423 is a homologue of the eukaryotic Rad52 protein. Following exposure to ionizing radiation, DR0423 expression is induced relative to an untreated control, and strains carrying a deletion of the DR0423 gene exhibit increased sensitivity to ionizing radiation. When recovering from ionizing-radiation-induced DNA damage in the absence of nutrients, wild-type D. radiodurans reassembles its genome while the mutant lacking DR0423 function does not. In vitro, the purified DR0423 protein binds to single-stranded DNA with an apparent affinity for 3' ends, and protects those ends from nuclease degradation. We propose that DR0423 is part of a DNA end-protection system that helps to preserve genome integrity following exposure to ionizing radiation. We designate the DR0423 protein as DNA damage response A protein.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
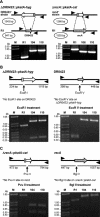
Figure 1. Verification of Gene Deletions
(A) Verification of ddrA and recA gene deletions by PCR analysis. Purified PCR fragments were amplified from the genomic DNA of strains R1, TNK104, TNK106, and TNK110 using primers that flank the coding sequences for ddrA and recA. Products were separated on a 0.8% agarose gel to establish whether the fragment size corresponded to the gene-replacement cassette. The left panel depicts the replacement of ddrA in TNK104 and TNK110. The right panel depicts the replacement of recA in TNK106 and TNK110. Expected sizes of the wild-type and mutant sequences are given in the figure above each image of the agarose gel. (B) Verification of the ddrA gene deletion by restriction analysis of purified PCR products. Purified PCR fragments were amplified from the genomic DNA of strains R1, TNK104, and TNK110, using primers that flank the coding sequences for ddrA. Products were restricted with EcoR1 (left panel) and EcoRV (right panel) to verify their identity. Products were separated on a 0.8% agarose gel to establish whether the restriction fragment corresponded with the expected sizes as illustrated in the figure above each image of the agarose gel. (C) Verification of the recA gene deletion by restriction analysis of purified PCR products. Purified PCR fragments were amplified from the genomic DNA of strains R1, TNK106, and TNK110, using primers that flank the coding sequences for recA. Products were restricted with PvuII (left panel) and BglII (right panel) to verify their identity. Products were separated on a 0.8% agarose gel to establish whether the restriction fragment corresponded with expected sizes as illustrated in the figure above each gel.
Figure 2. DNA Damage Sensitivity of D. radiodurans Cells Lacking DdrA Function
(A) Representative survival curves for D. radiodurans strain TNK104 ΔddrA (squares) and D. radiodurans R1 (circles) following exposure to γ radiation. Survival of strains; values are the mean ± standard deviation of three independent experiments; n = 9. (B) Representative survival curves for D. radiodurans strain TNK104 ΔddrA (squares) and D. radiodurans R1 (circles) following exposure to mitomycin C. Values are the mean ± standard deviation of three independent experiments; n = 9.
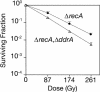
Figure 3. DdrA Functions in a RecA-Independent DNA Repair Process
Representative survival curves for D. radiodurans strains TNK106 ΔrecA (closed circles) and TNK110 ΔddrA ΔrecA (open triangles) following exposure to lower levels of γ radiation. All values are the mean ± standard deviation of three independent experiments; n = 9.
Figure 4. Genome Recovery in the Absence of Nutrients Depends on DdrA
(A) Pulsed-field gel electrophoresis analyses of D. radiodurans strain RI recovery over a 120 h time course in 10 mM MgSO4 following 5,000-Gy γ radiation. (B) Pulsed-field gel electrophoresis analyses of D. radiodurans strain TNK104 (ΔddrA) recovery following 5,000-Gy γ radiation.
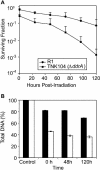
Figure 5. DdrA Protein Effects on In Vivo Survival and Genome Preservation following Exposure to Ionizing Radiation in the Absence of Nutrients
(A) Survival of D. radiodurans R1 and TNK104 cultures held in 10 mM MgSO4 for 120 h following exposure to 5,000-Gy γ radiation. Samples were obtained at 24-h intervals. All values are the mean ± standard deviation of three independent experiments; n = 9 (B) Changes in DNA content in cultures of R1 and TNK104 recovering from exposure to 5,000-Gy γ radiation in MgSO4. The DNA concentration at each time point is expressed as a percentage of that present in each strain prior to irradiation.
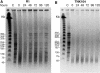
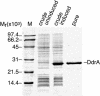
Figure 6. Purification of the DdrA Protein
The first lane contains molecular weight markers. The second and third lanes contain crude extracts from E. coli strain pEAW298 (DdrA overproducer) in which the ddrA gene is uninduced or induced, respectively. The final lane contains purified DdrA protein.
Figure 7. DdrA Protein Binds to Single-Stranded DNA with Free 3′ Ends
Four sets of EMSAs are presented, with the gels and electrophoresis conditions carefully matched. DNA substrate concentrations are 0.7 nM in each case, reported as total molecules. In each set, the first three lanes show the effects of the indicated concentration of DdrA protein. The fourth and fifth lanes are identical to the second and third lanes, respectively, except that they are treated with proteinase K to demonstrate that the DNA has not been altered. In set D, the sixth and seventh lanes are identical to the third lane (with 4 μM DdrA protein), except that they have been challenged with a 1,000-fold or 2,000-fold excess of unlabeled oligo with a 5′ extension, respectively. The unlabeled challenge oligo is the same as that used in reaction set C. (A) Single-stranded oligonucleotides (51 nt in length), labeled on the 5′ end. (B) 5′ end–labeled duplex DNA fragments (51 bp). (C) 5′ end–labeled oligonucleotide, with a self-complementary sequence leading to the formation of an 18-bp hairpin and a 15-nt 5′ single-stranded extension. (D) 3′ end–labeled oligonucleotide, with a self-complementary sequence leading to the formation of an 18-bp hairpin and a 16-nt 3′ single-stranded extension. The sequences of the single-stranded extensions in the oligos used in sets C and D are matched, except that an extra adenosine residue has been added to the oligo used in set D during the labeling process. Note that in set B, only the lower substrate band (unannealed oligonucleotides) is bound by DdrA, and the migration of the resulting complexes is identical to that shown in set A.
Figure 8. DdrA Protein Protects 3′ Ends from Degradation by Exonuclease I
(A) This set of reactions uses the labeled duplex DNA illustrated. The oligos annealed to form this DNA are 51 and 37 nt in length and pair so as to leave a 14-nt 3′ extension. The shorter DNA is 5′ end–labeled. The first lane contains unreacted DNA, showing both the annealed duplex and the unannealed single-stranded DNA. The second lane shows the DNA after treatment with 3 units of exonuclease I for 7 min in a 15-μl reaction mixture. Note that the duplex DNA in the upper band has been shortened by removal of the single-stranded extension. In lanes 3 and 4, the DdrA protein (4 μM) has been incubated with the DNA, without and with the 3 units of exonuclease I, respectively. The DNA is bound by DdrA and shifted to the top of the gel. The reactions shown in lanes 5 and 6 are identical to those in lanes 3 and 4, but with SDS and proteinase K added to disrupt the DdrA–DNA complexes and reveal that the DNA has been minimally affected by exonuclease I. The final lane shows another reaction of the DNA with 3 units of exonuclease I, in the presence of 4 μM bovine serum albumin. Exonuclease I degrades single-stranded DNA in the 3′ to 5′ direction. (B) The protein bound to the duplex DNA is DdrA. The reaction of lane 3 in (A) was scaled up and the protein–DNA complex excised from the gel as described in Materials and Methods. The protein in this complex was subjected to electrophoresis on an SDS-polyacrylamide gel, shown here (lane 3). The control lanes contained prestained protein standards (lane 1) and purified DdrA protein (lane 2). The gel-extracted protein comigrated with DdrA.
Similar articles
- Deinococcus radiodurans HD-Pnk, a Nucleic Acid End-Healing Enzyme, Abets Resistance to Killing by Ionizing Radiation and Mitomycin C.
Schmier BJ, Shuman S. Schmier BJ, et al. J Bacteriol. 2018 Aug 10;200(17):e00151-18. doi: 10.1128/JB.00151-18. Print 2018 Sep 1. J Bacteriol. 2018. PMID: 29891641 Free PMC article. - The stable, functional core of DdrA from Deinococcus radiodurans R1 does not restore radioresistance in vivo.
Harris DR, Ngo KV, Cox MM. Harris DR, et al. J Bacteriol. 2008 Oct;190(19):6475-82. doi: 10.1128/JB.01165-07. Epub 2008 Aug 1. J Bacteriol. 2008. PMID: 18676665 Free PMC article. - The IrrE protein of Deinococcus radiodurans R1 is a novel regulator of recA expression.
Earl AM, Mohundro MM, Mian IS, Battista JR. Earl AM, et al. J Bacteriol. 2002 Nov;184(22):6216-24. doi: 10.1128/JB.184.22.6216-6224.2002. J Bacteriol. 2002. PMID: 12399492 Free PMC article. - Gene regulation for the extreme resistance to ionizing radiation of Deinococcus radiodurans.
Wang W, Ma Y, He J, Qi H, Xiao F, He S. Wang W, et al. Gene. 2019 Oct 5;715:144008. doi: 10.1016/j.gene.2019.144008. Epub 2019 Jul 27. Gene. 2019. PMID: 31362038 Review. - Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics.
Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ. Makarova KS, et al. Microbiol Mol Biol Rev. 2001 Mar;65(1):44-79. doi: 10.1128/MMBR.65.1.44-79.2001. Microbiol Mol Biol Rev. 2001. PMID: 11238985 Free PMC article. Review.
Cited by
- Deinococcus as new chassis for industrial biotechnology: biology, physiology and tools.
Gerber E, Bernard R, Castang S, Chabot N, Coze F, Dreux-Zigha A, Hauser E, Hivin P, Joseph P, Lazarelli C, Letellier G, Olive J, Leonetti JP. Gerber E, et al. J Appl Microbiol. 2015 Jul;119(1):1-10. doi: 10.1111/jam.12808. Epub 2015 Apr 20. J Appl Microbiol. 2015. PMID: 25809882 Free PMC article. Review. - Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance.
Tanaka M, Earl AM, Howell HA, Park MJ, Eisen JA, Peterson SN, Battista JR. Tanaka M, et al. Genetics. 2004 Sep;168(1):21-33. doi: 10.1534/genetics.104.029249. Genetics. 2004. PMID: 15454524 Free PMC article. - Hypothetical proteins present during recovery phase of radiation resistant bacterium Deinococcus radiodurans are under purifying selection.
Das AD, Misra HS. Das AD, et al. J Mol Evol. 2013 Aug;77(1-2):31-42. doi: 10.1007/s00239-013-9577-9. Epub 2013 Aug 10. J Mol Evol. 2013. PMID: 23934623 - Radiation resistance in thermophiles: mechanisms and applications.
Ranawat P, Rawat S. Ranawat P, et al. World J Microbiol Biotechnol. 2017 Jun;33(6):112. doi: 10.1007/s11274-017-2279-5. Epub 2017 May 3. World J Microbiol Biotechnol. 2017. PMID: 28470425 Review. - DNA repair enzymes of the Antarctic Dry Valley metagenome.
Rzoska-Smith E, Stelzer R, Monterio M, Cary SC, Williamson A. Rzoska-Smith E, et al. Front Microbiol. 2023 Apr 14;14:1156817. doi: 10.3389/fmicb.2023.1156817. eCollection 2023. Front Microbiol. 2023. PMID: 37125210 Free PMC article.
References
- Anderson AW, Nordon HC, Cain RF, Parrish G, Duggan D. Studies on a radio-resistant micrococcus. I, Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 1956;10:575–578.
- Battista JR, Rainey FA. Family 1. Deinococcaceae. In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey's manual of systematic bacteriology, Volume 1, 2nd ed. New York: Springer; 2001. pp. 395–414.
- Battista JR, Earl AM, Park MJ. Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol. 1999;7:362–365. - PubMed
- Blondelet-Rouault MH, Weiser J, Lebrihi A, Branny P, Pernodet JL. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene. 1997;190:315–317. - PubMed
- Bonacossa de Almeida C, Coste G, Sommer S, Bailone A. Quantification of RecA protein in Deinococcus radiodurans reveals involvement of RecA, but not LexA, in its regulation. Mol Genet Genomics. 2002;268:28–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials