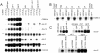Genetic analysis of pathways regulated by the von Hippel-Lindau tumor suppressor in Caenorhabditis elegans - PubMed (original) (raw)
Genetic analysis of pathways regulated by the von Hippel-Lindau tumor suppressor in Caenorhabditis elegans
Tammie Bishop et al. PLoS Biol. 2004 Oct.
Abstract
The von Hippel-Lindau (VHL) tumor suppressor functions as a ubiquitin ligase that mediates proteolytic inactivation of hydroxylated alpha subunits of hypoxia-inducible factor (HIF). Although studies of VHL-defective renal carcinoma cells suggest the existence of other VHL tumor suppressor pathways, dysregulation of the HIF transcriptional cascade has extensive effects that make it difficult to distinguish whether, and to what extent, observed abnormalities in these cells represent effects on pathways that are distinct from HIF. Here, we report on a genetic analysis of HIF-dependent and -independent effects of VHL inactivation by studying gene expression patterns in Caenorhabditis elegans. We show tight conservation of the HIF-1/VHL-1/EGL-9 hydroxylase pathway. However, persisting differential gene expression in hif-1 versus hif-1; vhl-1 double mutant worms clearly distinguished HIF-1-independent effects of VHL-1 inactivation. Genomic clustering, predicted functional similarities, and a common pattern of dysregulation in both vhl-1 worms and a set of mutants (dpy-18, let-268, gon-1, mig-17, and unc-6), with different defects in extracellular matrix formation, suggest that dysregulation of these genes reflects a discrete HIF-1-independent function of VHL-1 that is connected with extracellular matrix function.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
Figure 1. HIF-1–Dependent Effects of VHL-1 Inactivation
Representative RNase protection assays of genes that were differentially expressed in the vhl-1 versus wild-type microarray in (A) mixed-stage and (B) synchronized populations of worm. All genes are regulated by the VHL-1/HIF-1/EGL-9 pathway. (C) Regulation of nhr-57 mRNA in egl-9; vhl-1 worms and by egl-9 RNAi and DIP in vhl-1 worms. For RNAi experiments controls were L4440 vector alone (−) and C17G10.1, an irrelevant putative dioxygenase. F21C3.5 is a constitutively expressed gene used to control for RNA integrity. RNase protection assays were performed using worms cultured under normoxic conditions, unless otherwise indicated.
Figure 2. HIF-1–Independent Effects of VHL-1 Inactivation
RNase protection assays of genes that were differentially expressed in the hif-1; vhl-1 versus hif-1 microarrays in (A) mixed-stage and (B) synchronized populations of worm. The results confirm the existence of VHL-1–dependent, HIF-1–independent effects on gene expression.
Figure 3. Chromosomal Clustering of VHL-1–Dependent (HIF-1–Independent) Genes
(A) Chromosomal localization of VHL-1–dependent, HIF-1–independent genes. The positions of the genes from Table 4 are indicated by vertical ticks along the C. elegans chromosomes (shown to scale). Where two such genes are too close to be clearly resolved, the tick is marked by an asterisk. The single significant spatial clustering of VHL-1–dependent, HIF-1–independent genes is indicated by a red rectangle. The histogram under each chromosome shows the gene density (deeper bar, greater density) calculated as a sliding window of 100,000 bp moving with 10,000-bp increments along each chromosome. Dark blue indicates total annotated gene density, and light blue indicates the density of genes from the microarray that passed preliminary quality control. (B) Organization of the VHL-1–regulated (HIF-1–independent) gene cluster from Chromosome V. The relative positions and sizes of gene transcription units are shown to scale, with genes transcribed left to right above the horizontal line and right to left below the line. Names in black indicate genes that passed all selection criteria to be considered upregulated in hif-1; vhl-1 versus hif-1 worms (see Table 4). Genes with a mean >2.0-fold upregulation are indicated by green boxes, 1.5- to 2-fold are yellow, and <1.5-fold are red. Genes for which no data were obtained are shown as light grey.
Figure 4. Responses of VHL-1–Dependent, HIF-1–Independent Genes to egl-9 Inactivation, Hypoxia, and 2-Oxoglutarate Dioxygenase Inhibitors
RNase protection assays showing regulation of VHL-1–dependent, HIF-1–independent genes by (A) EGL-9 and hypoxia and (B) pharmacological inhibitors of 2-oxoglutarate dioxygenases: DIP and DMOG. None of the genes is regulated by EGL-9, but two genes (C01B4.7 and C01B4.8) show modest induction by hypoxia, DIP, and DMOG.
Figure 5. Sensitivity of VHL-1–Regulated Genes to Defects in Extracellular Matrix-Associated Proteins
RNase protection assays showing altered expression of VHL-1–regulated genes that are HIF-1 independent (upper six panels) and HIF-1 dependent (F22B5.4) in worms bearing mutations affecting (A) procollagen prolyl and lysyl hydroxylases and (B) other extracellular matrix-associated proteins. A common pattern of upregulation is observed in hif-1; vhl-1, vhl-1, dpy-18, let-268, gon-1, mig-17, and unc-6 worms but not other mutants. This contrasts with the HIF-1–dependent gene F22B5.4, which is upregulated in vhl-1 worms but none of the other mutants. (C) RNase protection assay for C01B4.9 illustrating DPY-18–mediated changes in expression that are independent of HIF-1.
Similar articles
- Identification of novel hypoxia dependent and independent target genes of the von Hippel-Lindau (VHL) tumour suppressor by mRNA differential expression profiling.
Wykoff CC, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Wykoff CC, et al. Oncogene. 2000 Dec 14;19(54):6297-305. doi: 10.1038/sj.onc.1204012. Oncogene. 2000. PMID: 11175344 - The Caenorhabditis elegans rhy-1 gene inhibits HIF-1 hypoxia-inducible factor activity in a negative feedback loop that does not include vhl-1.
Shen C, Shao Z, Powell-Coffman JA. Shen C, et al. Genetics. 2006 Nov;174(3):1205-14. doi: 10.1534/genetics.106.063594. Epub 2006 Sep 15. Genetics. 2006. PMID: 16980385 Free PMC article. - Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease.
Zatyka M, da Silva NF, Clifford SC, Morris MR, Wiesener MS, Eckardt KU, Houlston RS, Richards FM, Latif F, Maher ER. Zatyka M, et al. Cancer Res. 2002 Jul 1;62(13):3803-11. Cancer Res. 2002. PMID: 12097293 - Von Hippel-Lindau tumor suppressor protein and hypoxia-inducible factor in kidney cancer.
Maynard MA, Ohh M. Maynard MA, et al. Am J Nephrol. 2004 Jan-Feb;24(1):1-13. doi: 10.1159/000075346. Epub 2003 Dec 3. Am J Nephrol. 2004. PMID: 14654728 Review. - The von Hippel-Lindau tumor suppressor gene and kidney cancer.
Kaelin WG Jr. Kaelin WG Jr. Clin Cancer Res. 2004 Sep 15;10(18 Pt 2):6290S-5S. doi: 10.1158/1078-0432.CCR-sup-040025. Clin Cancer Res. 2004. PMID: 15448019 Review.
Cited by
- Flavin-containing monooxygenases in aging and disease: Emerging roles for ancient enzymes.
Rossner R, Kaeberlein M, Leiser SF. Rossner R, et al. J Biol Chem. 2017 Jul 7;292(27):11138-11146. doi: 10.1074/jbc.R117.779678. Epub 2017 May 17. J Biol Chem. 2017. PMID: 28515321 Free PMC article. Review. - Hif-1alpha regulates differentiation of limb bud mesenchyme and joint development.
Provot S, Zinyk D, Gunes Y, Kathri R, Le Q, Kronenberg HM, Johnson RS, Longaker MT, Giaccia AJ, Schipani E. Provot S, et al. J Cell Biol. 2007 May 7;177(3):451-64. doi: 10.1083/jcb.200612023. Epub 2007 Apr 30. J Cell Biol. 2007. PMID: 17470636 Free PMC article. - Hypoxia-inducible factor-dependent degeneration, failure, and malignant transformation of the heart in the absence of the von Hippel-Lindau protein.
Lei L, Mason S, Liu D, Huang Y, Marks C, Hickey R, Jovin IS, Pypaert M, Johnson RS, Giordano FJ. Lei L, et al. Mol Cell Biol. 2008 Jun;28(11):3790-803. doi: 10.1128/MCB.01580-07. Epub 2008 Feb 19. Mol Cell Biol. 2008. PMID: 18285456 Free PMC article. - NEDD8 acts as a 'molecular switch' defining the functional selectivity of VHL.
Russell RC, Ohh M. Russell RC, et al. EMBO Rep. 2008 May;9(5):486-91. doi: 10.1038/embor.2008.19. Epub 2008 Mar 7. EMBO Rep. 2008. PMID: 18323857 Free PMC article. - Renal cancer: oxygen meets metabolism.
Haase VH. Haase VH. Exp Cell Res. 2012 May 15;318(9):1057-67. doi: 10.1016/j.yexcr.2012.02.026. Epub 2012 Mar 3. Exp Cell Res. 2012. PMID: 22406000 Free PMC article. Review.
References
- Blelloch R, Kimble J. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans . Nature. 1999;399:586–590. - PubMed
- Blumenthal T, Gleason KS. Caenorhabditis elegans operons: Form and function. Nat Rev Genet. 2003;4:112–120. - PubMed
- Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. - PubMed
- Camenisch G, Stroka DM, Gassmann M, Wenger RH. Attenuation of HIF-1 DNA-binding activity limits hypoxia-inducible endothelin-1 expression. Pflugers Arch. 2001;443:240–249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials