Population history and natural selection shape patterns of genetic variation in 132 genes - PubMed (original) (raw)
Population history and natural selection shape patterns of genetic variation in 132 genes
Joshua M Akey et al. PLoS Biol. 2004 Oct.
Abstract
Identifying regions of the human genome that have been targets of natural selection will provide important insights into human evolutionary history and may facilitate the identification of complex disease genes. Although the signature that natural selection imparts on DNA sequence variation is difficult to disentangle from the effects of neutral processes such as population demographic history, selective and demographic forces can be distinguished by analyzing multiple loci dispersed throughout the genome. We studied the molecular evolution of 132 genes by comprehensively resequencing them in 24 African-Americans and 23 European-Americans. We developed a rigorous computational approach for taking into account multiple hypothesis tests and demographic history and found that while many apparent selective events can instead be explained by demography, there is also strong evidence for positive or balancing selection at eight genes in the European-American population, but none in the African-American population. Our results suggest that the migration of modern humans out of Africa into new environments was accompanied by genetic adaptations to emergent selective forces. In addition, a region containing four contiguous genes on Chromosome 7 showed striking evidence of a recent selective sweep in European-Americans. More generally, our results have important implications for mapping genes underlying complex human diseases.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
Figure 1. Scatter Plot of Neutrality Test Statistics in European- and African-Americans
Genes that are nominally significant (p < 0.05) in European-Americans (EA), African-Americans (AA), or both populations are denoted by red, blue, and green circles, respectively. Genes that are not significant are shown as black dots. Two-sided tests were used for Tajima's D, Fu and Li's D*, and Fu and Li's F*, and a one-sided test was used for Fay and Wu's H.
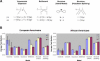
Figure 2. Summary of the Four Demographic Models Considered in Each Population
(A) Schematic diagram of each demographic model and its associated parameters (see Materials and Methods for details). Parameter values that match the observed data most closely for European-Americans (EA) and African-Americans (AA) are shown below the diagrams. (B) Average and 95% confidence intervals of Tajima's D (blue bars), Fu and Li's D* (red bars), and Fu and Li's F* (pale yellow bars) for the observed data and each demographic model (using the parameters that most closely match the empirical data). Results from the standard neutral model (Constant) are also shown.
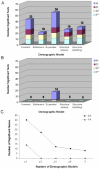
Figure 3. The Influence of Demographic History on Tests of Selection
(A and B) The significance of observed values of Tajima's D (red), Fu and Li's D* (pale yellow), Fu and Li's F* (pale blue), and Fay and Wu's H (dark blue) were reassessed for each best-fit demographic model in European-Americans (A) and African-Americans (B). Results from the standard neutral model (Constant) are shown for comparison. The number of significant genes for each demographic model is noted above each category in (A) and (B). For example, there were a total of 19 significant test statistics across all four tests of neutrality assuming a bottleneck model for Europeans, which define ten unique genes. Therefore, each gene is supported by approximately two (19/10) tests of neutrality. (C) The distribution of the number of significant genes across the five demographic models in European-Americans and African-Americans. For example, in European-Americans, 40 genes were significant in at least one of the demographic models, and 27 genes were significant in at least two of the demographic models.
Figure 4. A Strong Signature of Positive Selection Spanning 115 kb on Chromosome 7q
(A–D) Exons for EPHB6, TRPV6, TRPV5, and KEL are shown as gray vertical lines. A dashed black line indicates the boundary between EPHB6 and TRPV6 exons, which are approximately 1 kb apart. Transcriptional orientation is indicated by the arrows below exon positions. SNPs found in European-Americans and African-Americans are shown below. Noncoding, synonymous, and nonsynonymous SNPs are denoted as black, blue, and red vertical bars, respectively. The positions of three nonsynonymous SNPs in TRPV6 are shown with asterisks. For each of the resulting nonsynonymous amino acid changes, the most frequent amino acid in European-Americans is given first. The frequency of derived alleles, PD (B), sliding window plots of Tajima's D (C), and nucleotide diversity, π (D), are shown across the entire region. Gaps in the sliding window plots indicate positions where sequence data were not obtained. In (B–D), European- and African-American data are shown in red and black, respectively. (E) The distribution of FST across the 115-kb region. The average FST for all SNPs across the 132 genes is shown as a dashed red line. The dashed green line indicates the threshold for significantly (p < 0.01) large values of FST, determined by coalescent simulations.
Similar articles
- Disentangling the effects of demography and selection in human history.
Stajich JE, Hahn MW. Stajich JE, et al. Mol Biol Evol. 2005 Jan;22(1):63-73. doi: 10.1093/molbev/msh252. Epub 2004 Sep 8. Mol Biol Evol. 2005. PMID: 15356276 - Assessing the evolutionary impact of amino acid mutations in the human genome.
Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, White TJ, Nielsen R, Clark AG, Bustamante CD. Boyko AR, et al. PLoS Genet. 2008 May 30;4(5):e1000083. doi: 10.1371/journal.pgen.1000083. PLoS Genet. 2008. PMID: 18516229 Free PMC article. - Darwinian and demographic forces affecting human protein coding genes.
Nielsen R, Hubisz MJ, Hellmann I, Torgerson D, Andrés AM, Albrechtsen A, Gutenkunst R, Adams MD, Cargill M, Boyko A, Indap A, Bustamante CD, Clark AG. Nielsen R, et al. Genome Res. 2009 May;19(5):838-49. doi: 10.1101/gr.088336.108. Epub 2009 Mar 11. Genome Res. 2009. PMID: 19279335 Free PMC article. - Genetic variation and adaptation in Africa: implications for human evolution and disease.
Gomez F, Hirbo J, Tishkoff SA. Gomez F, et al. Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a008524. doi: 10.1101/cshperspect.a008524. Cold Spring Harb Perspect Biol. 2014. PMID: 24984772 Free PMC article. Review. - Genome-wide scans for loci under selection in humans.
Ronald J, Akey JM. Ronald J, et al. Hum Genomics. 2005 Jun;2(2):113-25. doi: 10.1186/1479-7364-2-2-113. Hum Genomics. 2005. PMID: 16004726 Free PMC article. Review.
Cited by
- A cross-population extended haplotype-based homozygosity score test to detect positive selection in genome-wide scans.
Zhong M, Zhang Y, Lange K, Fan R. Zhong M, et al. Stat Interface. 2011;4(1):51-63. doi: 10.4310/SII.2011.v4.n1.a6. Stat Interface. 2011. PMID: 26097641 Free PMC article. - A high resolution genome-wide scan for significant selective sweeps: an application to pooled sequence data in laying chickens.
Qanbari S, Strom TM, Haberer G, Weigend S, Gheyas AA, Turner F, Burt DW, Preisinger R, Gianola D, Simianer H. Qanbari S, et al. PLoS One. 2012;7(11):e49525. doi: 10.1371/journal.pone.0049525. Epub 2012 Nov 29. PLoS One. 2012. PMID: 23209582 Free PMC article. - Reduced food intake and body weight in mice deficient for the G protein-coupled receptor GPR82.
Engel KM, Schröck K, Teupser D, Holdt LM, Tönjes A, Kern M, Dietrich K, Kovacs P, Krügel U, Scheidt HA, Schiller J, Huster D, Brockmann GA, Augustin M, Thiery J, Blüher M, Stumvoll M, Schöneberg T, Schulz A. Engel KM, et al. PLoS One. 2011;6(12):e29400. doi: 10.1371/journal.pone.0029400. Epub 2011 Dec 28. PLoS One. 2011. PMID: 22216272 Free PMC article. - The case for selection at CCR5-Delta32.
Sabeti PC, Walsh E, Schaffner SF, Varilly P, Fry B, Hutcheson HB, Cullen M, Mikkelsen TS, Roy J, Patterson N, Cooper R, Reich D, Altshuler D, O'Brien S, Lander ES. Sabeti PC, et al. PLoS Biol. 2005 Nov;3(11):e378. doi: 10.1371/journal.pbio.0030378. Epub 2005 Nov 1. PLoS Biol. 2005. PMID: 16248677 Free PMC article. - Genetic Ancestry Estimates within Dutch Family Units and Across Genotyping Arrays: Insights from Empirical Analysis Using Two Estimation Methods.
Beck JJ, Ahmed T, Finnicum CT, Zwinderman K, Ehli EA, Boomsma DI, Hottenga JJ. Beck JJ, et al. Genes (Basel). 2023 Jul 22;14(7):1497. doi: 10.3390/genes14071497. Genes (Basel). 2023. PMID: 37510400 Free PMC article.
References
- Akey JM, Zhang K, Xiong M, Jin L. The effect of single nucleotide polymorphism identification strategies on estimates of linkage disequilibrium. Mol Biol Evol. 2003;20:232–242. - PubMed
- Andolfatto P. Adaptive hitchhiking effects on genome variability. Curr Opin Genet Dev. 2001;11:635–641. - PubMed
- Cavalli-Sforza LL. Population structure and human evolution. Proc R Soc Lond B Biol Sci. 1966;164:362–379. - PubMed
Publication types
MeSH terms
Grants and funding
- U01 HL066642/HL/NHLBI NIH HHS/United States
- R37 MH059520/MH/NIMH NIH HHS/United States
- MH59520/MH/NIMH NIH HHS/United States
- U01 HL69757/HL/NHLBI NIH HHS/United States
- HL66682/HL/NHLBI NIH HHS/United States
- HL66642/HL/NHLBI NIH HHS/United States
- R01 MH059520/MH/NIMH NIH HHS/United States
- U01 HL069757/HL/NHLBI NIH HHS/United States
- U01 HL066682/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials