A novel class of mammalian-specific tailless retropseudogenes - PubMed (original) (raw)
. 2004 Oct;14(10A):1911-5.
doi: 10.1101/gr.2720104. Epub 2004 Sep 13.
Affiliations
- PMID: 15364902
- PMCID: PMC524414
- DOI: 10.1101/gr.2720104
A novel class of mammalian-specific tailless retropseudogenes
Jürgen Schmitz et al. Genome Res. 2004 Oct.
Abstract
In addition to their central function in protein biosynthesis, tRNAs also play a pervasive role in genome evolution and architecture because of their extensive ability to serve as templates for retroposition. Close to half of the human genome consists of discernible transposable elements, a vast majority of which are derived from RNA via reverse transcription and genomic integration. Apart from the presence of direct repeats (DRs) that flank the integrated sequence of retroposons, genomic integrations are usually marked by an oligo(A) tail. Here, we describe a novel class of retroposons that lack A-tails and are therefore termed tailless retropseudogenes. Analysis of approximately 2500 tRNA-related young tailless retropseudogene sequences revealed that they comprise processed and unprocessed (pre-)tRNAs, 3'-truncated in their loop regions, or truncated tRNA-derived SINE RNAs. Surprisingly, their mostly nonrandom integration is dependent on the priming of reverse transcription at sites determined by their 3'-terminal 2-18 nucleotides and completely independent from oligoadenylation of the template RNA. Thus, tailless retropseudogenes point to a novel, variant mechanism for the biogenesis of retrosequences.
Figures
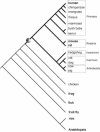
Figure 1
Taxonomic distribution of tailless retropseudogenes based on GenBank searches. Tailless retropseudogenes were retrieved exclusively from mammalian representatives (bold lines) indicating a mammalian-specific distribution with the presumed origin marked by a dot. Species represented in GenBank with their entire genome sequences are shown in bold letters. Branches of the phylogenetic tree are not drawn to scale.
Figure 2
Genomic integration mechanism of tRNA-derived tailless retropseudogenes. (A) RNA break point (arrow) and priming site (boxed nucleotides) in an example in which a fragment of pre-tRNACys serves as a tailless retropseudogene template. Tailless retropseudogene-specific substitutions differing from the presumed tRNA precursor template are encircled. Nucleotides representing the remainder of the tRNA precursor, but not part of the tailless retropseudogenes, are depicted as “thin” letters. (B) The genomic integration site with the consensus sequence TT/WX1-17 (in this particular case, TT/WX6) is shown above the bracket. The RNA priming site is boxed, the direction of reverse transcription is indicated by an arrow. The presumed single-stranded, staggered end is shown by a double-headed arrow, part of which (seven nucleotides in this case) acts as a DNA primer for reverse transcription. (C) Sequence of chromosomal locus after integration of the tailless retropseudogenes. Direct repeats (DR) are indicated by arrows. Note that the priming site (within the boxed tailless retropseudogenes) is part of the DR.
Figure 3
Precursor and cleavage point of a tailless retropseudogene from an intron-containing tRNA. In an example where human pre-tRNATyr serves as a precursor for a tailless retropseudogene, the RNA breakpoint 3′ to a purine residue (arrow) is located in the intron (lower case letters). The priming site, also located in the intron, is boxed. The boxed nucleotides at the 5′-end represent unprocessed nucleotides that are present in the integrated tailless retropseudogene. Nucleotides representing the remainder of the tRNA precursor, but not part of the tailless retropseudogene, are depicted by “thin” letters.
Figure 4
Tailless retropseudogene presence/absence alignments for primates. In the alignments of orthologous loci for M1-M9 markers (one or several with tailless retropseudogenes and one or more orthologous loci without tailless retropseudogenes) all direct repeats are boxed and priming sites are shaded. The nonorthologous, most closely related tRNA species is given in the last line of each of the nine alignments. Hyphens represent gaps in the sequences. Homo sapiens (Hs); Pan troglodytes (Pt); Gorilla gorilla (Gg); Pongo pygmaeus (Pp); Hylobates lar (Hl) for Hominoidea; Macaca mulatta (Mm) for Cercopithecoidea; Lagothrix lagotricha (Ll) for Platyrrhini; Lemur catta (Lc) for Strepsirrhini. Accession nos. or internal reference numbers are shown in bold.
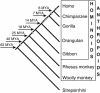
Figure 5
Timeframes of integration of primate tailless retropseudogenes defined by presence/absence analyses. Dots on the lineage leading to human mark the nine established single-integration events. Species splitting times are adapted from Goodman and Page (Goodman et al. 1998).
Similar articles
- Ancient traces of tailless retropseudogenes in therian genomes.
Noll A, Raabe CA, Churakov G, Brosius J, Schmitz J. Noll A, et al. Genome Biol Evol. 2015 Feb 26;7(3):889-900. doi: 10.1093/gbe/evv040. Genome Biol Evol. 2015. PMID: 25724209 Free PMC article. - Similar target site selection occurs in integration of plant and mammalian retroposons.
Tatout C, Lavie L, Deragon JM. Tatout C, et al. J Mol Evol. 1998 Oct;47(4):463-70. doi: 10.1007/pl00006403. J Mol Evol. 1998. PMID: 9767691 - MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation.
Smit AF, Riggs AD. Smit AF, et al. Nucleic Acids Res. 1995 Jan 11;23(1):98-102. doi: 10.1093/nar/23.1.98. Nucleic Acids Res. 1995. PMID: 7870595 Free PMC article. - RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements.
Brosius J. Brosius J. Gene. 1999 Sep 30;238(1):115-34. doi: 10.1016/s0378-1119(99)00227-9. Gene. 1999. PMID: 10570990 Review. - Retrotransposal integration of mobile genetic elements in human diseases.
Miki Y. Miki Y. J Hum Genet. 1998;43(2):77-84. doi: 10.1007/s100380050045. J Hum Genet. 1998. PMID: 9621510 Review.
Cited by
- RNA-Mediated Gene Duplication and Retroposons: Retrogenes, LINEs, SINEs, and Sequence Specificity.
Ohshima K. Ohshima K. Int J Evol Biol. 2013;2013:424726. doi: 10.1155/2013/424726. Epub 2013 Aug 1. Int J Evol Biol. 2013. PMID: 23984183 Free PMC article. - Sequence evidence in the archaeal genomes that tRNAs emerged through the combination of ancestral genes as 5' and 3' tRNA halves.
Fujishima K, Sugahara J, Tomita M, Kanai A. Fujishima K, et al. PLoS One. 2008 Feb 20;3(2):e1622. doi: 10.1371/journal.pone.0001622. PLoS One. 2008. PMID: 18286179 Free PMC article. - Large-scale discovery of insertion hotspots and preferential integration sites of human transposed elements.
Levy A, Schwartz S, Ast G. Levy A, et al. Nucleic Acids Res. 2010 Mar;38(5):1515-30. doi: 10.1093/nar/gkp1134. Epub 2009 Dec 14. Nucleic Acids Res. 2010. PMID: 20008508 Free PMC article. - Gene discovery at the human T-cell receptor alpha/delta locus.
Haynes MR, Wu GE. Haynes MR, et al. Immunogenetics. 2007 Feb;59(2):109-21. doi: 10.1007/s00251-006-0165-7. Epub 2006 Dec 13. Immunogenetics. 2007. PMID: 17165047 - Different integration site structures between L1 protein-mediated retrotransposition in cis and retrotransposition in trans.
Kojima KK. Kojima KK. Mob DNA. 2010 Jul 8;1(1):17. doi: 10.1186/1759-8753-1-17. Mob DNA. 2010. PMID: 20615209 Free PMC article.
References
- Brosius, J. 1999a. Transmutation of tRNA over time. Nat. Genet. 22: 8-9. - PubMed
- ____. 1999b. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238: 115-134. - PubMed
- ____. 2001. tRNAs in the spotlight during protein biosynthesis. Trends Biochem. Sci. 26: 653-656. - PubMed
- Cost, G.J. and Boeke, J.D. 1998. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37: 18081-18093. - PubMed
WEB SITE REFERENCES
- http://zmbe2.unimuenster.de/expath/addmat/tailless_retropseudogenes1.htm; supplementary data, including sequences and additional priming sites.
- http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker; A.F.A. Smit and P. Green, RepeatMasker.
- http://rna.wustl.edu/GtRDB/Hs/Hs-align.html; tRNAscan-SE online tRNA Analysis.
- http://genome.ucsc.edu/cgi-bin/hgGateway; UCSC Genome Browser Gateway.
- http://genome.ucsc.edu/cgi-bin/hgBlat; UCSC BLAT Search Genome.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources