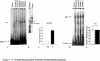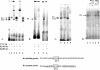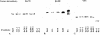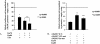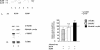The Ku protein complex interacts with YY1, is up-regulated in human heart failure, and represses alpha myosin heavy-chain gene expression - PubMed (original) (raw)
The Ku protein complex interacts with YY1, is up-regulated in human heart failure, and represses alpha myosin heavy-chain gene expression
Carmen C Sucharov et al. Mol Cell Biol. 2004 Oct.
Abstract
Human heart failure is accompanied by repression of genes such as alpha myosin heavy chain (alphaMyHC) and SERCA2A and the induction of fetal genes such as betaMyHC and atrial natriuretic factor. It seems likely that changes in MyHC isoforms contribute to the poor contractility seen in heart failure, because small changes in isoform composition can have a major effect on the contractility of cardiac myocytes and the heart. Our laboratory has recently shown that YY1 protein levels are increased in human heart failure and that YY1 represses the activity of the human alphaMyHC promoter. We have now identified a region of the alphaMyHC promoter that binds a factor whose expression is increased sixfold in failing human hearts. Through peptide mass spectrometry, we identified this binding activity to be a heterodimer of Ku70 and Ku80. Expression of Ku represses the human alphaMyHC promoter in neonatal rat ventricular myocytes. Moreover, overexpression of Ku70/80 decreases alphaMyHC mRNA expression and increases skeletal alpha-actin. Interestingly, YY1 interacts with Ku70 and Ku80 in HeLa cells. Together, YY1, Ku70, and Ku80 repress the alphaMyHC promoter to an extent that is greater than that with YY1 or Ku70/80 alone. Our results suggest that Ku is an important factor in the repression of the human alphaMyHC promoter during heart failure.
Figures
FIG. 1.
Failing human heart extracts have elevated αMyHC DNA binding activity. (A) The −370/−350 region of the human αMyHC promoter was incubated with nonfailing and failing heart extracts and assayed by EMSA. (B) The same probe was incubated with failing heart and HeLa cell extracts. (C) The binding complexes were quantified and plotted on a graph. (D) EMSA with a probe containing an Sp1 binding site was incubated with nonfailing and failing heart extracts. (E) The binding complexes were quantified and plotted on a graph.
FIG. 2.
Purification of the Ku70/80 complex. (A) EMSA of the eluted fractions from the Dynabead purification (see Materials and Methods). Lane 1, 200 mM KCl elution; lane 2, 300 mM KCl elution; lane 3, 1 M KCl elution; lane 4, HeLa cell nuclear extract. (B) Cross-linking experiments using the eluted fractions from the Dynabead purification. Lane 1, flowthrough; lane 2, 1 M KCl elution; lane 3, 200 mM KCl elution. (C) Denaturing polyacrylamide gel of the 1 M elution fraction. Lane 1, 1 M elution fraction; lane 2, 1 M elution of the fraction containing beads only; lane 3, flowthrough. The arrows represent the five bands that were analyzed by mass spectrometry. Arrows 1 and 2 indicate Ku70 and Ku80, respectively.
FIG. 3.
The Ku70/80 complex binds to the αMyHC promoter in a specific manner. (A) EMSA using the wild-type probe incubated with HeLa cell nuclear extract (lane 1), HeLa cell nuclear extract plus YY1 Ab (lane 2), HeLa cell nuclear extract plus Ku70 Ab (lane 3), purified Ku proteins (lane 4), purified Ku proteins plus Ku70 Ab (lane 5), or purified Ku proteins plus YY1 Ab (lane 6). (B) EMSA using the wild-type probe incubated with failing human heart extract (lanes 1 and 3), failing heart nuclear extract plus Ku70 Ab (lane 2), or failing heart nuclear extract plus Ku80 Ab (lane 4). (C) EMSA using probes containing mutations in the YY1 binding site (lane 2), Ku binding site (lane 3), or Ku and YY1 binding site (lane 4) incubated with HeLa cell nuclear extract. (D) EMSA using the YY1 mutant probe incubated with HeLa cell nuclear extract, with increasing amounts of the Ku wild-type (W.T.) competitor (lanes 2 and 3) and mutant (Mut) competitor (lanes 4 and 5).
FIG. 4.
Ku recognizes and binds to the −370/−350 region of the αMyHC promoter independently of DNA ends. (A) Effects of Exo III on linear and circular probes (see Materials and Methods). (B) EMSA of the linear and circular probes incubated with HeLa cell nuclear extract and purified Ku70/80 proteins with or without Exo III. (C) Schematic representation of the linear and circular probes. The arrow on each probe corresponds to the αMyHC fragment that contains the Ku binding site.
FIG. 5.
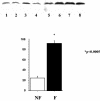
Ku70 protein levels are increased in failing heart extracts, as shown in Western blotting experiments using Ku70 Ab. Lanes 1 to 4, extract from nonfailing human hearts; lanes 5 to 8, extract from failing human hearts. The relative amount of Ku70 protein is plotted in the graph.
FIG. 6.
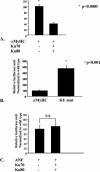
The Ku70/80 complex represses the activity of the αMyHC promoter in NRVM. (A) Cotransfection of Ku70/80 repressed the activity of the αMyHC promoter. (B) Mutations in the Ku binding site resulted in a fivefold up-regulation of the αMyHC promoter. (C) Cotransfection of Ku70/80 did not change the activity of the ANF promoter.
FIG. 7.
YY1 interacts with Ku70 and Ku80. HeLa cell extracts were incubated with Abs indicated at the bottom of each panel without (-1) or with (-2) ethidium bromide, and Western blotting experiments were performed with the indicated Abs. (A) Western blot of Ku70; (B) Western blot of Ku80; (C) Western blot of YY1.
FIG. 8.
YY1 and Ku70/80 together increase the repression of αMyHC promoter activity. (A) Effects of YY1 and Ku70/80 in NRVM on the activity of the αMyHC promoter. The results are normalized to the αMyHC promoter cotransfected with an empty vector. (B) Cotransfection of Ku70/80 and an αMyHC promoter construct containing the YY1 or the Ku binding sites mutated in NRVM resulted in lower repression levels. The wild-type and mutant constructs were normalized to 100%. The activity of the mutant construct was higher than that of the wild type (37) (Fig. 6).
FIG. 9.
Ku70 and Ku80 repress endogenous αMyHC gene expression. (A) Overexpression of Ku70 and Ku80 together but neither one alone results in increased levels of both proteins, as shown by Western blotting of NRVM infected with adenovirus expressing Ku70 and/or Ku80. Lanes 1 to 3, Ku70 Ab; lanes 4 to 6, Ku80 Ab. Lanes 1 and 4, nuclear extract from NRVM infected with a control adenovirus; lane 2, nuclear extract from NRVM infected with adenovirus expressing Ku70; lanes 3 and 6, nuclear extract from NRVM infected with adenovirus expressing Ku70 and Ku80; lane 5, nuclear extract from NRVM infected with adenovirus expressing Ku80. (B) Representative RPA. Lane 1, infection with control virus; lane 2, infection with Ku70 and Ku80 viruses. (C) Endogenous αMyHC mRNA levels are repressed by overexpression of Ku70 and Ku80. RPA experiments were done, and the results are plotted.
Similar articles
- Yin Yang 1 is increased in human heart failure and represses the activity of the human alpha-myosin heavy chain promoter.
Sucharov CC, Mariner P, Long C, Bristow M, Leinwand L. Sucharov CC, et al. J Biol Chem. 2003 Aug 15;278(33):31233-9. doi: 10.1074/jbc.M301917200. Epub 2003 May 16. J Biol Chem. 2003. PMID: 12754214 - Yin Yang 1 represses alpha-myosin heavy chain gene expression in pathologic cardiac hypertrophy.
Mariner PD, Luckey SW, Long CS, Sucharov CC, Leinwand LA. Mariner PD, et al. Biochem Biophys Res Commun. 2005 Jan 7;326(1):79-86. doi: 10.1016/j.bbrc.2004.11.008. Biochem Biophys Res Commun. 2005. PMID: 15567155 - The repressor which binds the -75 GATA motif of the GPB promoter contains Ku70 as the DNA binding subunit.
Camara-Clayette V, Thomas D, Rahuel C, Barbey R, Cartron JP, Bertrand O. Camara-Clayette V, et al. Nucleic Acids Res. 1999 Apr 1;27(7):1656-63. doi: 10.1093/nar/27.7.1656. Nucleic Acids Res. 1999. PMID: 10075997 Free PMC article. - YY1 represses vitamin D receptor-mediated 25-hydroxyvitamin D(3)24-hydroxylase transcription: relief of repression by CREB-binding protein.
Raval-Pandya M, Dhawan P, Barletta F, Christakos S. Raval-Pandya M, et al. Mol Endocrinol. 2001 Jun;15(6):1035-46. doi: 10.1210/mend.15.6.0651. Mol Endocrinol. 2001. PMID: 11376120 - IL-1 beta increases abundance and activity of the negative transcriptional regulator yin yang-1 (YY1) in neonatal rat cardiac myocytes.
Patten M, Wang W, Aminololama-Shakeri S, Burson M, Long CS. Patten M, et al. J Mol Cell Cardiol. 2000 Jul;32(7):1341-52. doi: 10.1006/jmcc.2000.1169. J Mol Cell Cardiol. 2000. PMID: 10860774
Cited by
- NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes.
Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J, Carathers M, Davuluri R, Guttridge DC. Wang H, et al. Mol Cell Biol. 2007 Jun;27(12):4374-87. doi: 10.1128/MCB.02020-06. Epub 2007 Apr 16. Mol Cell Biol. 2007. PMID: 17438126 Free PMC article. - Reversible epigenetic modifications of the two cardiac myosin heavy chain genes during changes in expression.
Pandya K, Pulli B, Bultman S, Smithies O. Pandya K, et al. Gene Expr. 2010;15(2):51-9. doi: 10.3727/105221611x12973615737505. Gene Expr. 2010. PMID: 21526716 Free PMC article. - Factors controlling cardiac myosin-isoform shift during hypertrophy and heart failure.
Gupta MP. Gupta MP. J Mol Cell Cardiol. 2007 Oct;43(4):388-403. doi: 10.1016/j.yjmcc.2007.07.045. Epub 2007 Jul 21. J Mol Cell Cardiol. 2007. PMID: 17720186 Free PMC article. Review. - Pervasive Chromatin-RNA Binding Protein Interactions Enable RNA-Based Regulation of Transcription.
Xiao R, Chen JY, Liang Z, Luo D, Chen G, Lu ZJ, Chen Y, Zhou B, Li H, Du X, Yang Y, San M, Wei X, Liu W, Lécuyer E, Graveley BR, Yeo GW, Burge CB, Zhang MQ, Zhou Y, Fu XD. Xiao R, et al. Cell. 2019 Jun 27;178(1):107-121.e18. doi: 10.1016/j.cell.2019.06.001. Cell. 2019. PMID: 31251911 Free PMC article. - Identification of Ku70 Domain-Specific Interactors Using BioID2.
Abbasi S, Schild-Poulter C. Abbasi S, et al. Cells. 2021 Mar 14;10(3):646. doi: 10.3390/cells10030646. Cells. 2021. PMID: 33799447 Free PMC article.
References
- Abraham, W. T., E. M. Gilbert, B. D. Lowes, W. A. Minobe, P. Larrabee, R. L. Roden, D. Dutcher, J. Sederberg, J. A. Lindenfeld, E. E. Wolfel, S. F. Shakar, D. Ferguson, K. Volkman, J. V. Linseman, R. A. Quaife, A. D. Robertson, and M. R. Bristow. 2002. Coordinate changes in myosin heavy chain isoform gene expression are selectively associated with alterations in dilated cardiomyopathy phenotype. Mol. Med. 8:750-760. - PMC - PubMed
- Albert, H., E. C. Dale, E. Lee, and D. W. Ow. 1995. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7:649-659. - PubMed
- Bertinato, J., J. J. Tomlinson, C. Schild-Poulter, and R. J. Hache. 2003. Evidence implicating Ku antigen as a structural factor in RNA polymerase II-mediated transcription. Gene 302:53-64. - PubMed
- Bliss, T. M., and D. P. Lane. 1997. Ku selectively transfers between DNA molecules with homologous ends. J. Biol. Chem. 272:5765-5773. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials