Stress granule assembly is mediated by prion-like aggregation of TIA-1 - PubMed (original) (raw)
Stress granule assembly is mediated by prion-like aggregation of TIA-1
Natalie Gilks et al. Mol Biol Cell. 2004 Dec.
Abstract
TIA-1 is an RNA binding protein that promotes the assembly of stress granules (SGs), discrete cytoplasmic inclusions into which stalled translation initiation complexes are dynamically recruited in cells subjected to environmental stress. The RNA recognition motifs of TIA-1 are linked to a glutamine-rich prion-related domain (PRD). Truncation mutants lacking the PRD domain do not induce spontaneous SGs and are not recruited to arsenite-induced SGs, whereas the PRD forms aggregates that are recruited to SGs in low-level-expressing cells but prevent SG assembly in high-level-expressing cells. The PRD of TIA-1 exhibits many characteristics of prions: concentration-dependent aggregation that is inhibited by the molecular chaperone heat shock protein (HSP)70; resistance to protease digestion; sequestration of HSP27, HSP40, and HSP70; and induction of HSP70, a feedback regulator of PRD disaggregation. Substitution of the PRD with the aggregation domain of a yeast prion, SUP35-NM, reconstitutes SG assembly, confirming that a prion domain can mediate the assembly of SGs. Mouse embryomic fibroblasts (MEFs) lacking TIA-1 exhibit impaired ability to form SGs, although they exhibit normal phosphorylation of eukaryotic initiation factor (eIF)2alpha in response to arsenite. Our results reveal that prion-like aggregation of TIA-1 regulates SG formation downstream of eIF2alpha phosphorylation in response to stress.
Figures
Figure 1.
Prion-like domains in TIA-1, TIAR, Sup 35, and human prion protein. (A) Amino acid composition of the prion-like domains of TIA-1 and TIAR, the N-terminal prion domain of Sup35, and the aggregation domain of human prion protein. Y, G, N, and Q residues are shaded, and glutamine residues (Q) are underlined. (B) Schematic representation of TIA-1 and Sup35 constructs and full-length proteins. The PRD constructs in pMT2-HA and pSRα-HA-GFP vectors begin at methionine 230. The pSRα-HA-PRD initiates at methionine 219. H indicates the N-terminal HA tag.
Figure 2.
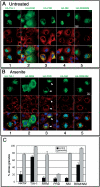
Replacement of the PRD of TIA-1 with the NM domain of yeast Sup35 restores normal function to the RRM truncation mutant. (A and B) Immunofluorescent micrographs of transiently expressed HA-tagged TIA-1, RRM, PRD, SUP35-NM, and RRM/NM in pSRα vector expressed in COS7 cells. Cells were untreated (A) or treated with 0.5 mM arsenite for 45 min (B) to induce stress granule formation before fixation and staining for HA (green), eIF3 (red), and DNA (blue). Bars, 10 μm. (C) Quantification of the percent of transfectants containing stress granules, with and without arsenite treatment. At least 100 transfectants were scored for each construct, and results averaged from three independent experiments.
Figure 3.
Aggregated forms of TIA-1 and PRD in COS7 cells. (A and B) Light micrographs of COS7 cells expressing recombinant HA-TIA-1 (A) or HA-PRD (B) stained for HA. (C–H) Transmission electron micrographs of COS7 TIA-1 (untagged) transfectants (C–E) or untagged PRD transfectants (F–H). (C) Arrowheads indicate nuclear and cytoplasmic inclusions of TIA-1 not observed in vector transfectants. N, nucleus; C, cytoplasm; M, mitochondria. (D) Enlarged view of cytoplasmic inclusions of TIA-1. Arrowheads indicate ribosome-like particles surrounding the cytoplasmic aggregates. (E) Immunoelectron micrograph of TIA-1 transfectant, labeled with anti-TIA-1 antibody and visualized using a gold-conjugated anti-mouse Ig. Arrowheads indicate nuclear inclusions of TIA-1. (F) Transmission electron micrograph of PRD transfectant. Arrowheads indicate cytoplasmic aggregates of recombinant PRD. (G) Enlarged view of cytoplasmic aggregates. Arrowheads indicate ribosome-like particles that are not associated with the PRD aggregates. (H) Immunoelectron micrograph of PRD transfectant stained for TIA-1 by using immunogold. Arrowheads indicate cytoplasmic aggregates of PRD, which stain less intensely with lead citrate than do TIA-1 aggregates but are strongly stained with anti-TIA-1 immunogold.
Figure 4.
GFP-PRD progressively forms insoluble aggregates that prevent SG assembly. (A) COS7 cells expressing GFP-tagged versions of TIA-1 or PRD shown at 24 and 48 h after transfection. GFP-TIA-1 (green) expression induces spontaneous SG assembly (arrows, confirmed by eIF3 staining, red), whereas GFP-PRD does not induce SGs (green, arrowheads) and becomes predominantly cytoplasmic at later times (48 h). (B) COS7 transfectants as in A, treated with 0.5 mM sodium arsenite for 45 min Bar, 25 um. (C) Quantification of the percentage of GFP-TIA-1 (white bars) or GFP-PRD (black bars) transfectants that exhibit SGs. Transfected cells were cultured for 24, 36, or 48 h and either untreated or exposed to arsenite (0.5 μM) for 45 min. Data are expressed as the percentage of transfected cells with eIF3+ SGs. Data shown are typical of three independent experiments. (D) Western blot of GFP-PRD and GFP-TIA-1 protein in detergent-soluble and detergent-insoluble fractions prepared from cells harvested at the indicated times. Asterisk indicates the mobility of GFP-TIA-1; arrowhead indicates mobility of GFP-PRD. (E) Densitometric quantification of the data shown in D. White bars, GFP-TIA-1; black bars, GFP-PRD.
Figure 5.
Protease resistance of GFP-PRD. (A) GFP-PRD is more resistant to proteolysis that GFP-TIA-1. COS7 cell lysates containing GFP-TIA-1 or GFP-PRD were prepared from 48-h transfectants and then incubated with the indicated concentrations of protease K for the times indicated at room temperature. Top, ponceau red staining of total protein. Middle, blot probed with anti-TIA-1 antibody against the C-terminal region. Bottom, blot probed for endogenous HSP40. (B) HA-TIA-1 and HA-PRD lysates, treated as in A. (C) Protease resistance of GFP-PRD increases with time. COS7 cells expressing GFP-TIA-1 or GFP-PRD were cultured for 24 or 48 h, lysed, and sonicated, and then treated with the indicated concentrations of protease K for 30 min at room temperature. Top, total protein. Middle, digestion of GFP-TIA-1. Bottom, digestion of GFP-PRD; blots probed with the antibody against the TIA-1 C terminus.
Figure 6.
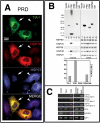
PRD aggregation induces HSP70 expression and sequesters HSP27. (A) Immunofluorescence micrographs of HA-PRD transfectants at 46 h, stained for TIA-1 (green), endogenous HSP70 (red), and endogenous HSP27 (blue). (B) Solubility of different HA-tagged proteins. COS7 transfectants were fractionated into insoluble and soluble fractions as described, resolved by PAGE, and probed with antibodies against HA, HSP90, HSP70, and HSP27. Top, blot probed with anti-HA; bottom, blot probed for HSP70, HSP40, and HSP27 as indicated. Graph shows quantification of percentage of insoluble recombinant protein. The HA-NM construct was not detected by Western blot, and its solubility could not be determined. (C) RT-PCR analysis of RNA from pMT2 vector and TIA-1 and PRD transfectants. Activation of the unfolded protein response causes the splicing of XBP1 mRNA to its smaller, active form. Primers used for this PCR reaction spanned the spliced region, creating a smaller band upon activation of the unfolded protein response.
Figure 7.
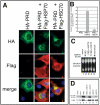
Overexpression of exogenous HSP70, but not HSC70, reverses the effects of the PRD on reporter gene expression. (A) Immunofluorescence micrographs of transfectants expressing the HA-PRD truncation mutant cotransfected with pcDNA3 vector, FLAG-tagged HSP70, or FLAG-tagged HSC70, stained for HA (green), FLAG (red), or DNA (blue). (B) Quantification of effects of vector, HSP70, or HSC70 on nuclear localization of HA-PRD in COS7. Cells were stained with anti-HA and anti-FLAG antibodies and manually scored for nuclear localization of HA-PRD. The results are the average of four independent experiments. (C) Northern blot analysis of COS7 transfectants coexpressing HA-PRD with pcDNA3 vector, HSP70, or HSC70. The HSP70 3′UTR probe recognized only the endogenous levels of HSP70 mRNA in all samples, not the exogenous HSP70 mRNA. RNA extracted from untransfected, heat shocked COS7 cells (42°C for 90 min) was used as a positive control. (D) Immunoblot showing effects of different constructs on reporter gene expression. Equal amounts of protein were loaded, and expression of the indicated constructs was verified by reprobing the same blot (our unpublished data).
Figure 8.
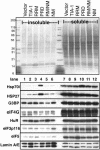
Solubility of SG components in cells expressing mutant forms of TIA-1. COS transfectants were fractionated into soluble and insoluble fractions as described in Figure 6B and probed for additional SG markers. Top, ponceau red staining showing total protein. Numbers indicate mobility of molecular weight markers in kilodaltons. Blots were probed for HSP70, HSP27, G3BP, eIF4G, HuR, eIF3p116, eIF5, and lamin A/E (as a loading control), as indicated.
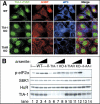
Figure 9.
TIA-1 KO MEFs exhibit impaired SG assembly. (A) MEFs from wild-type, TIA-1 –/– or TIAR –/– MEFs were exposed to 0.5 mM arsenite for 30 min and then fixed and stained for TIA (green, using a mixture of anti-TIA-1 and TIAR antibodies), G3BP (red), and eIF3 (blue). Bar, 10 μm. (B) MEFs from wild-type, TIA-1 –/–, TIAR –/–, or S51A mutant cells (as a nonphosphorylated control) were untreated (lanes 1, 5, 9, and 13) or treated for 30 min with 0.125 mM arsenite (lanes 2, 6, and 10), 0.5 mM arsenite (lanes 3, 7, and 11), or 0.25 mM arsenite (lanes 4, 8, 12, and 14). Blots were probed for phospho-eIF2α, p70S6 kinase (as a loading control), HuR, and TIA-1.
Similar articles
- Chandipura Virus Forms Cytoplasmic Inclusion Bodies through Phase Separation and Proviral Association of Cellular Protein Kinase R and Stress Granule Protein TIA-1.
Sarkar S, Ganguly S, Ganguly NK, Sarkar DP, Sharma NR. Sarkar S, et al. Viruses. 2024 Jun 26;16(7):1027. doi: 10.3390/v16071027. Viruses. 2024. PMID: 39066190 Free PMC article. - RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules.
Kedersha NL, Gupta M, Li W, Miller I, Anderson P. Kedersha NL, et al. J Cell Biol. 1999 Dec 27;147(7):1431-42. doi: 10.1083/jcb.147.7.1431. J Cell Biol. 1999. PMID: 10613902 Free PMC article. - Stress granules: sites of mRNA triage that regulate mRNA stability and translatability.
Kedersha N, Anderson P. Kedersha N, et al. Biochem Soc Trans. 2002 Nov;30(Pt 6):963-9. doi: 10.1042/bst0300963. Biochem Soc Trans. 2002. PMID: 12440955 Review. - Fibril-induced glutamine-/asparagine-rich prions recruit stress granule proteins in mammalian cells.
Riemschoss K, Arndt V, Bolognesi B, von Eisenhart-Rothe P, Liu S, Buravlova O, Duernberger Y, Paulsen L, Hornberger A, Hossinger A, Lorenzo-Gotor N, Hogl S, Müller SA, Tartaglia G, Lichtenthaler SF, Vorberg IM. Riemschoss K, et al. Life Sci Alliance. 2019 Jul 2;2(4):e201800280. doi: 10.26508/lsa.201800280. Print 2019 Aug. Life Sci Alliance. 2019. PMID: 31266883 Free PMC article. - RNA recognition and stress granule formation by TIA proteins.
Waris S, Wilce MC, Wilce JA. Waris S, et al. Int J Mol Sci. 2014 Dec 16;15(12):23377-88. doi: 10.3390/ijms151223377. Int J Mol Sci. 2014. PMID: 25522169 Free PMC article. Review.
Cited by
- Stress granules in cancer: Adaptive dynamics and therapeutic implications.
Jia Y, Jia R, Dai Z, Zhou J, Ruan J, Chng W, Cai Z, Zhang X. Jia Y, et al. iScience. 2024 Jun 22;27(8):110359. doi: 10.1016/j.isci.2024.110359. eCollection 2024 Aug 16. iScience. 2024. PMID: 39100690 Free PMC article. Review. - Axonopathy Underlying Amyotrophic Lateral Sclerosis: Unraveling Complex Pathways and Therapeutic Insights.
Luan T, Li Q, Huang Z, Feng Y, Xu D, Zhou Y, Hu Y, Wang T. Luan T, et al. Neurosci Bull. 2024 Aug 4. doi: 10.1007/s12264-024-01267-2. Online ahead of print. Neurosci Bull. 2024. PMID: 39097850 Review. - Chandipura Virus Forms Cytoplasmic Inclusion Bodies through Phase Separation and Proviral Association of Cellular Protein Kinase R and Stress Granule Protein TIA-1.
Sarkar S, Ganguly S, Ganguly NK, Sarkar DP, Sharma NR. Sarkar S, et al. Viruses. 2024 Jun 26;16(7):1027. doi: 10.3390/v16071027. Viruses. 2024. PMID: 39066190 Free PMC article. - Ataxin-2: a powerful RNA-binding protein.
Li L, Wang M, Huang L, Zheng X, Wang L, Miao H. Li L, et al. Discov Oncol. 2024 Jul 22;15(1):298. doi: 10.1007/s12672-024-01158-y. Discov Oncol. 2024. PMID: 39039334 Free PMC article. Review. - UBAP2L contributes to formation of P-bodies and modulates their association with stress granules.
Riggs CL, Kedersha N, Amarsanaa M, Zubair SN, Ivanov P, Anderson P. Riggs CL, et al. J Cell Biol. 2024 Oct 7;223(10):e202307146. doi: 10.1083/jcb.202307146. Epub 2024 Jul 15. J Cell Biol. 2024. PMID: 39007803 Free PMC article.
References
- Aguzzi, A., and Polymenidou, M. (2004). Mammalian prion biology: one century of evolving concepts. Cell 116, 313–327. - PubMed
- Akakura, S., Yoshida, M., Yoneda, Y., and Horinouchi, S. (2001). A role for Hsc70 in regulating nucleocytoplasmic transport of a temperature-sensitive p53 (p53Val-135). J. Biol. Chem. 276, 14649–14657. - PubMed
- Anderson, P., and Kedersha, N. (2002a). Stressful initiations. J. Cell Sci. 115, 3227–3234. - PubMed
- Busch, A., Engemann, S., Lurz, R., Okazawa, H., Lehrach, H., and Wanker, E.E. (2003). Mutant huntingtin promotes the fibrillogenesis of wild-type huntingtin: a potential mechanism for loss of huntingtin function in Huntington's disease. J. Biol. Chem. 278, 41452–41461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 AI033600/AI/NIAID NIH HHS/United States
- R01 AI033600/AI/NIAID NIH HHS/United States
- AI33600/AI/NIAID NIH HHS/United States
- R01 AI050167/AI/NIAID NIH HHS/United States
- AI50167/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous