Bifidobacterium longum requires a fructokinase (Frk; ATP:D-fructose 6-phosphotransferase, EC 2.7.1.4) for fructose catabolism - PubMed (original) (raw)
Bifidobacterium longum requires a fructokinase (Frk; ATP:D-fructose 6-phosphotransferase, EC 2.7.1.4) for fructose catabolism
Cristina I Caescu et al. J Bacteriol. 2004 Oct.
Abstract
Although the ability of Bifidobacterium spp. to grow on fructose as a unique carbon source has been demonstrated, the enzyme(s) needed to incorporate fructose into a catabolic pathway has hitherto not been defined. This work demonstrates that intracellular fructose is metabolized via the fructose-6-P phosphoketolase pathway and suggests that a fructokinase (Frk; EC 2.7.1.4) is the enzyme that is necessary and sufficient for the assimilation of fructose into this catabolic route in Bifidobacterium longum. The B. longum A10C fructokinase-encoding gene (frk) was expressed in Escherichia coli from a pET28 vector with an attached N-terminal histidine tag. The expressed enzyme was purified by affinity chromatography on a Co(2+)-based column, and the pH and temperature optima were determined. A biochemical analysis revealed that Frk displays the same affinity for fructose and ATP (Km(fructose) = 0.739 +/- 0.18 mM and Km(ATP) = 0.756 +/- 0.08 mM), is highly specific for D-fructose, and is inhibited by an excess of ATP (>12 mM). It was also found that frk is inducible by fructose and is subject to glucose-mediated repression. Consequently, this work presents the first characterization at the molecular and biochemical level of a fructokinase from a gram-positive bacterium that is highly specific for D-fructose.
Figures
FIG. 1.
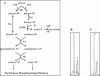
(A) Intermediates and enzymes of the F6PPK pathway for
d
-glucose catabolism in the genus Bifidobacterium. HK, hexokinase (EC 2.7.1.1); PGI, glucose-phosphate isomerase (EC 5.3.1.9); F6PPK, F6P phosphoketolase (EC 4.1.2. 9.); AK, acetokinase (EC 2.7.2.1); TA, transaldolase (EC 2.2.1.2); TK, transketolase (EC 2.2.1.1). HPAEC-PAD profiles of
d
-glucose (B) and
d
-fructose (C) fermentation by a crude extract of B. longum A10C are shown. Peaks of glucose-6-phosphate (1), F6P (2), and sedoheptulose-7-phosphate (3) are indicated.
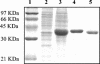
FIG. 2.
Purification of the fructokinase Frk. Samples (1 μg of protein) were separated in a sodium dodecyl sulfate-10% polyacrylamide gel and stained with Coomassie brilliant blue R-250. Lane 1, molecular mass standard; lane 2, negative control [a crude extract of E. coli BL21 (DE3) containing pET28a(+)]; lane 3, crude extract of IPTG-induced cells of E. coli BL21 (DE3) containing pFrk; lane 4, eluate with sonication buffer containing 150 mM imidazole from cobalt-based affinity column; lane 5, purified Frk after elimination of the six-His tag by thrombin cleavage.
FIG. 3.
TLC analysis of Frk carbohydrate substrate specificity.
d
-Fructose (lane 1), F6P (lane 2),
d
-mannose (lane 4), mannose-6-phosphate (lane 6),
d
-glucose (lane 7), glucose-6-phosphate (lane 8), and sucrose (lane 12) were used as standards (5 μg/lane). The phosphorylated carbohydrates formed during the incubation of Frk with
d
-fructose (lane 3),
d
-mannose (lane 5),
d
-glucose (lane 9),
l
-arabinose (lane 10),
d
-xylose (lane 11), and sucrose (lane 13) are shown.
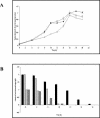
FIG. 4.
Profile of carbohydrate utilization by B. longum A10C throughout the growth curve. (A) B. longum A10C growth curve. Cells were cultivated in modified Garches medium supplemented with glucose (diamonds), fructose (squares), or an equimolar mix of the two hexoses (triangles) for the indicated times. (B) Amounts of nonfermented sugars, as measured by HPAEC-PAD. Black bars,
d
-glucose; gray bars,
d
-fructose; checked bars,
d
-glucose in mixed GF medium; hatched bars,
d
-fructose in mixed GF medium.
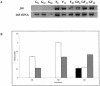
FIG. 5.
RT-PCR analysis of frk transcripts produced in B. longum A10C cells grown on different carbohydrate sources. (A) Primers specific for frk sequence and 16S rDNA were used to amplify fragments by RT-PCR. (B) Calculated ratios of frk to 16S rDNA (RT-PCR band intensities of specific products). Total RNAs were isolated from B. longum A10C cultivated for 8, 12, and 20 h on modified Garches medium containing 2 g of the following monosaccharides liter−1: glucose (G; black bars), fructose (F; white bars), or an equimolar mix of glucose and fructose (GF; gray bars).
FIG. 6.
Genetic context of frk locus in two B. longum strains (NCC2705 and DJ010A) and genotype analysis of the fructose-negative strain DSM20219. (A) EcoRV restriction map of a 9.2-kb fragment of the chromosome of B. longum NCC2705 surrounding the frk locus. The additional 1,119-bp noncoding sequence separating frk from the downstream ORF on the DJO10A genome and the additional EcoRV4 site are indicated with large parentheses. (B) Analysis by agarose gel electrophoresis of amplicons generated by PCRs performed with fructose-positive A10C (+) and fructose-negative DSM20219 (−) genomic DNAs. The following sets of primers were designed to amplify DNA fragments located around the frk locus: 1-2 (861 bp), 3-4 (768 bp), _glk_ATG-_glk_TAA (908 bp), _frk_ATG-_frk_TAA (894 bp), and 7-8 (784 bp). (C) (Left) Southern blot of EcoRV-digested DNA from the A10C (Fru+) and DSM20219 (Fru−) strains hybridized with probes 3 and 4. (Right) Southern blot of EcoRV-digested DNA from the A10C (Fru+) and DSM20219 (Fru−) strains hybridized with the frk probe. Arrows indicate the specific bands detected with probes amplified from B. longum A10C genomic DNA. On both gels, the central lane corresponds to DNA ladder X (Roche).
FIG. 7.
(A) HPAEC-PAD analysis of fructose degradation by B. longum DSM20219 crude extract. (B) HPAEC-PAD analysis of fructose degradation by B. longum DSM20219 crude extract supplemented with 0.1 U of purified Frk. Peaks corresponding to
d
-fructose (1), glucose-6-phosphate (2), F6P (3), and sedoheptulose-7-phosphate (4) are indicated.
Similar articles
- Cloning, sequencing, and expression of the Zymomonas mobilis fructokinase gene and structural comparison of the enzyme with other hexose kinases.
Zembrzuski B, Chilco P, Liu XL, Liu J, Conway T, Scopes R. Zembrzuski B, et al. J Bacteriol. 1992 Jun;174(11):3455-60. doi: 10.1128/jb.174.11.3455-3460.1992. J Bacteriol. 1992. PMID: 1317376 Free PMC article. - Identification and biochemical characterization of the fructokinase gene family in Arabidopsis thaliana.
Riggs JW, Cavales PC, Chapiro SM, Callis J. Riggs JW, et al. BMC Plant Biol. 2017 Apr 26;17(1):83. doi: 10.1186/s12870-017-1031-5. BMC Plant Biol. 2017. PMID: 28441933 Free PMC article. - Characterisation of glutamine fructose-6-phosphate amidotransferase (EC 2.6.1.16) and N-acetylglucosamine metabolism in Bifidobacterium.
Foley S, Stolarczyk E, Mouni F, Brassart C, Vidal O, Aïssi E, Bouquelet S, Krzewinski F. Foley S, et al. Arch Microbiol. 2008 Feb;189(2):157-67. doi: 10.1007/s00203-007-0307-9. Epub 2007 Oct 18. Arch Microbiol. 2008. PMID: 17943273 - Plant Fructokinases: Evolutionary, Developmental, and Metabolic Aspects in Sink Tissues.
Stein O, Granot D. Stein O, et al. Front Plant Sci. 2018 Mar 16;9:339. doi: 10.3389/fpls.2018.00339. eCollection 2018. Front Plant Sci. 2018. PMID: 29616058 Free PMC article. Review. - The anhydrofructose pathway and its possible role in stress response and signaling.
Yu S, Fiskesund R. Yu S, et al. Biochim Biophys Acta. 2006 Sep;1760(9):1314-22. doi: 10.1016/j.bbagen.2006.05.007. Epub 2006 Jun 2. Biochim Biophys Acta. 2006. PMID: 16822618 Review.
Cited by
- Fructose uptake in Bifidobacterium longum NCC2705 is mediated by an ATP-binding cassette transporter.
Wei X, Guo Y, Shao C, Sun Z, Zhurina D, Liu D, Liu W, Zou D, Jiang Z, Wang X, Zhao J, Shang W, Li X, Liao X, Huang L, Riedel CU, Yuan J. Wei X, et al. J Biol Chem. 2012 Jan 2;287(1):357-367. doi: 10.1074/jbc.M111.266213. Epub 2011 Nov 18. J Biol Chem. 2012. PMID: 22102285 Free PMC article. - Mechanism of high-mannose N-glycan breakdown and metabolism by Bifidobacterium longum.
Cordeiro RL, Santos CR, Domingues MN, Lima TB, Pirolla RAS, Morais MAB, Colombari FM, Miyamoto RY, Persinoti GF, Borges AC, de Farias MA, Stoffel F, Li C, Gozzo FC, van Heel M, Guerin ME, Sundberg EJ, Wang LX, Portugal RV, Giuseppe PO, Murakami MT. Cordeiro RL, et al. Nat Chem Biol. 2023 Feb;19(2):218-229. doi: 10.1038/s41589-022-01202-4. Epub 2022 Nov 28. Nat Chem Biol. 2023. PMID: 36443572 Free PMC article. - Molecular and biochemical characterization of Entamoeba histolytica fructokinase.
Matt J, Duchêne M. Matt J, et al. Parasitol Res. 2015 May;114(5):1939-47. doi: 10.1007/s00436-015-4383-5. Epub 2015 Feb 21. Parasitol Res. 2015. PMID: 25700717 Free PMC article. - Kinetic analysis of bifidobacterial metabolism reveals a minor role for succinic acid in the regeneration of NAD+ through its growth-associated production.
Van der Meulen R, Adriany T, Verbrugghe K, De Vuyst L. Van der Meulen R, et al. Appl Environ Microbiol. 2006 Aug;72(8):5204-10. doi: 10.1128/AEM.00146-06. Appl Environ Microbiol. 2006. PMID: 16885266 Free PMC article. - Milk glycan metabolism by intestinal bifidobacteria: insights from comparative genomics.
Arzamasov AA, Osterman AL. Arzamasov AA, et al. Crit Rev Biochem Mol Biol. 2022 Oct-Dec;57(5-6):562-584. doi: 10.1080/10409238.2023.2182272. Epub 2023 Mar 3. Crit Rev Biochem Mol Biol. 2022. PMID: 36866565 Free PMC article. Review.
References
- Angell, S., E. Schwarz, and M. J. Bibb. 1992. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol. Microbiol. 6:2833-2844. - PubMed
- Aulkemeyer, P., R. Ebner, G. Heilenmann, K. Jahreis, K. Schmid, S. Wrieden, and J. W. Lengeler. 1991. Molecular analysis of two fructokinases involved in sucrose metabolism of enteric bacteria. Mol. Microbiol. 5:2913-2922. - PubMed
- Berghauser, J. 1975. A reactive arginine in adenylate kinase. Biochim. Biophys. Acta 397:370-376. - PubMed
- Bockmann, J., H. Heuel, and J. W. Lengeler. 1992. Characterization of a chromosomally encoded, non-PTS metabolic pathway for sucrose utilization in Escherichia coli EC3132. Mol. Gen. Genet. 235:22-32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous