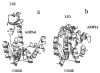Escherichia coli adenylate kinase dynamics: comparison of elastic network model modes with mode-coupling (15)N-NMR relaxation data - PubMed (original) (raw)
Comparative Study
. 2004 Nov 15;57(3):468-80.
doi: 10.1002/prot.20226.
Affiliations
- PMID: 15382240
- PMCID: PMC1752299
- DOI: 10.1002/prot.20226
Comparative Study
Escherichia coli adenylate kinase dynamics: comparison of elastic network model modes with mode-coupling (15)N-NMR relaxation data
N Alpay Temiz et al. Proteins. 2004.
Abstract
The dynamics of adenylate kinase of Escherichia coli (AKeco) and its complex with the inhibitor AP(5)A, are characterized by correlating the theoretical results obtained with the Gaussian Network Model (GNM) and the anisotropic network model (ANM) with the order parameters and correlation times obtained with Slowly Relaxing Local Structure (SRLS) analysis of (15)N-NMR relaxation data. The AMPbd and LID domains of AKeco execute in solution large amplitude motions associated with the catalytic reaction Mg(+2)*ATP + AMP --> Mg(+2)*ADP + ADP. Two sets of correlation times and order parameters were determined by NMR/SRLS for AKeco, attributed to slow (nanoseconds) motions with correlation time tau( perpendicular) and low order parameters, and fast (picoseconds) motions with correlation time tau( parallel) and high order parameters. The structural connotation of these patterns is examined herein by subjecting AKeco and AKeco*AP(5)A to GNM analysis, which yields the dynamic spectrum in terms of slow and fast modes. The low/high NMR order parameters correlate with the slow/fast modes of the backbone elucidated with GNM. Likewise, tau( parallel) and tau( perpendicular) are associated with fast and slow GNM modes, respectively. Catalysis-related domain motion of AMPbd and LID in AKeco, occurring per NMR with correlation time tau( perpendicular), is associated with the first and second collective slow (global) GNM modes. The ANM-predicted deformations of the unliganded enzyme conform to the functional reconfiguration induced by ligand-binding, indicating the structural disposition (or potential) of the enzyme to bind its substrates. It is shown that NMR/SRLS and GNM/ANM analyses can be advantageously synthesized to provide insights into the molecular mechanisms that control biological function.
(c) 2004 Wiley-Liss, Inc.
Figures
Fig. 1
Ribbon diagrams of the crystal structures of (a) AKeco and (b) AKeco in complex with the two-substrate-mimic inhibitor AP5A. The figures were drawn with the program Molscript using the PDB coordinate files 4ake for AKeco and 1 ake (complex II) for AKeco*AP5A. The AMP binding domain, AMPbd, comprises helices α2 and α3, the domain LID comprises the strands β5-β8 and the intervening loops, and the CORE domain comprises the remaining part of the polypeptide chain.
Fig. 2
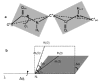
a: Schematic of the virtual bond of the GNM theory. The chain is made of consecutive peptide planes comprising the atoms . The bond vector Ni-Hi makes an angle εi with the virtual bond, Ii, that connects the atoms .b: Diagram showing the angular change Δαi in the orientationmi(t) of the bond Ni-Hi, induced by the torsional rotation, Δφi, undergone by the virtual bond, Ii. Δαi and Δφi are related by eq 14, assuming that the angle ε between Ii andmi(t) is fixed.
Fig. 3
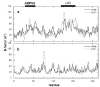
Experimental, (---) and GNM-predicted (—) B-factors for AKeco (a) and AKeco*AP5A (b).
Fig. 4
a: NMR order parameters obtained with MF analysis (open circles) and theoretical GNM order parameters (solid curve), as a function of residue number for AKeco.b: NMR order parameters obtained with SRLS analysis (open circles) and theoretical GNM order parameters calculated from the slowest N/4 modes (solid curve) and the N/4 fastest modes (dashed curve) as a function of residue number for AKeco.c: Counterpart of Figure a for AKeco*AP5A with the NMR/MF data from reference . d: Counterpart of b for AKeco*AP5A with the NMR/SRLS data from reference . (S02)2 and S2 were obtained by fitting the experimental 15N T1, T2, and15N- NOE data of the two enzyme forms obtained at 303K and at 14.1/18.8 T with SRLS, and MF, respectively, using τm = 15.1 ns for AKeco and τm= 11.6 ns for AKeco*AP5A.
Fig. 5
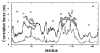
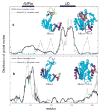
Best-fit local motion correlation time component,τ⊥;, obtained by fitting the experimental 15N T1, T2, and15N- NOE data of AKeco obtained at 303K, and at 14.1/18.8 T (open circles). GNM correlation times τiGNM (eq 12) obtained for AKeco from the five slowest modes (solid curve) and all the modes (dashed curve). The abscissa represents residue number.
Fig. 6
First slowest global GNM mode shapes for AKeco (—) and AKeco*AP5A (- - -) (a), and second slowest global GNM mode shapes for AKeco (—) and AKeco*AP5A (- - -) (b).Insets: The ribbon diagrams of AKeco and AKeco*AP5A color-coded cyan-blue-red-yellow-green in order of increasing mobility. The black boxes depict the LID and AMPbd domains. The blue dots denote residues associated by crystallographic studies with hinges.
Fig. 7
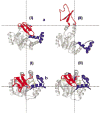
Conformations I and II between which AKeco (a) and AKeco*AP5A (b) fluctuate based on the first global mode according to ANM analysis. The LID and AMPbd domains are colored red and blue, respectively, and the inhibitor is colored yellow.
Fig. 8
Distribution of mean-square fluctuations determined with GNM for the AKmt solution structure and the AKeco*AP5A crystal structure as a function of the AKmt sequence.
Similar articles
- Domain flexibility in ligand-free and inhibitor-bound Escherichia coli adenylate kinase based on a mode-coupling analysis of 15N spin relaxation.
Shapiro YE, Kahana E, Tugarinov V, Liang Z, Freed JH, Meirovitch E. Shapiro YE, et al. Biochemistry. 2002 May 21;41(20):6271-81. doi: 10.1021/bi012132q. Biochemistry. 2002. PMID: 12009888 - A novel view of domain flexibility in E. coli adenylate kinase based on structural mode-coupling (15)N NMR relaxation.
Tugarinov V, Shapiro YE, Liang Z, Freed JH, Meirovitch E. Tugarinov V, et al. J Mol Biol. 2002 Jan 11;315(2):155-70. doi: 10.1006/jmbi.2001.5231. J Mol Biol. 2002. PMID: 11779236 - Activation energy of catalysis-related domain motion in E. coli adenylate kinase.
Shapiro YE, Meirovitch E. Shapiro YE, et al. J Phys Chem B. 2006 Jun 15;110(23):11519-24. doi: 10.1021/jp060282a. J Phys Chem B. 2006. PMID: 16771428 - Conformational heterogeneity within the LID domain mediates substrate binding to Escherichia coli adenylate kinase: function follows fluctuations.
Schrank TP, Wrabl JO, Hilser VJ. Schrank TP, et al. Top Curr Chem. 2013;337:95-121. doi: 10.1007/128_2012_410. Top Curr Chem. 2013. PMID: 23543318 Free PMC article. Review. - Induced-fit movements in adenylate kinases.
Schulz GE. Schulz GE. Faraday Discuss. 1992;(93):85-93. doi: 10.1039/fd9929300085. Faraday Discuss. 1992. PMID: 1290942 Review.
Cited by
- The interplay of structure and dynamics: insights from a survey of HIV-1 reverse transcriptase crystal structures.
Seckler JM, Leioatts N, Miao H, Grossfield A. Seckler JM, et al. Proteins. 2013 Oct;81(10):1792-801. doi: 10.1002/prot.24325. Epub 2013 Aug 16. Proteins. 2013. PMID: 23720322 Free PMC article. - iGNM: a database of protein functional motions based on Gaussian Network Model.
Yang LW, Liu X, Jursa CJ, Holliman M, Rader AJ, Karimi HA, Bahar I. Yang LW, et al. Bioinformatics. 2005 Jul 1;21(13):2978-87. doi: 10.1093/bioinformatics/bti469. Epub 2005 Apr 28. Bioinformatics. 2005. PMID: 15860562 Free PMC article. - Conformational transitions of adenylate kinase: switching by cracking.
Whitford PC, Miyashita O, Levy Y, Onuchic JN. Whitford PC, et al. J Mol Biol. 2007 Mar 9;366(5):1661-71. doi: 10.1016/j.jmb.2006.11.085. Epub 2006 Dec 5. J Mol Biol. 2007. PMID: 17217965 Free PMC article. - A conformational intermediate in glutamate receptor activation.
Lau AY, Salazar H, Blachowicz L, Ghisi V, Plested AJ, Roux B. Lau AY, et al. Neuron. 2013 Aug 7;79(3):492-503. doi: 10.1016/j.neuron.2013.06.003. Neuron. 2013. PMID: 23931998 Free PMC article. - Predicting the complex structure and functional motions of the outer membrane transporter and signal transducer FecA.
Sen TZ, Kloster M, Jernigan RL, Kolinski A, Bujnicki JM, Kloczkowski A. Sen TZ, et al. Biophys J. 2008 Apr 1;94(7):2482-91. doi: 10.1529/biophysj.107.116046. Epub 2008 Jan 4. Biophys J. 2008. PMID: 18178655 Free PMC article.
References
- Ishima R, Torchia DA. Protein dynamics from NMR. Nat Struct Biol. 2000;7:740–743. - PubMed
- Kay LE. Protein dynamics from NMR. Nat Struct Biol. 1998;5:513–517. - PubMed
- Palmer AG. NMR probes of molecular dynamics: Overview and comparison with other techniques. Annu Rev Biophys and Biomol Struct. 2001;30:129–155. - PubMed
- Peng JW, Wagner G. James TL, Oppenheimer NJ, editor. Methods Enzymol. New York: Academic Press;1994. p 563–595.
- Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity J Am Chem Soc. 1982;104:4546–4559.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases