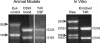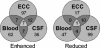Microarray analysis of pneumococcal gene expression during invasive disease - PubMed (original) (raw)
Microarray analysis of pneumococcal gene expression during invasive disease
Carlos J Orihuela et al. Infect Immun. 2004 Oct.
Abstract
Streptococcus pneumoniae is a leading cause of invasive bacterial disease. This is the first study to examine the expression of S. pneumoniae genes in vivo by using whole-genome microarrays available from The Institute for Genomic Research. Total RNA was collected from pneumococci isolated from infected blood, infected cerebrospinal fluid, and bacteria attached to a pharyngeal epithelial cell line in vitro. Microarray analysis of pneumococcal genes expressed in these models identified body site-specific patterns of expression for virulence factors, transporters, transcription factors, translation-associated proteins, metabolism, and genes with unknown function. Contributions to virulence predicted for several unknown genes with enhanced expression in vivo were confirmed by insertion duplication mutagenesis and challenge of mice with the mutants. Finally, we cross-referenced our results with previous studies that used signature-tagged mutagenesis and differential fluorescence induction to identify genes that are potentially required by a broad range of pneumococcal strains for invasive disease.
Figures
FIG. 1.
RNA collected from S. pneumoniae in vivo and in vitro. After isolation of pneumococcal RNA from infected blood, CSF, and bacteria adherent to the Detroit pharyngeal epithelial cell line, RNA was visualized on a 1% TBE gel to confirm purity and assess degradation. Bacterial RNA (open arrowheads) and eukaryotic RNA (shaded arrowheads) are indicated. Bacterial RNA isolated from the ECC model was enriched after collection of the initial pellet (raw) to remove contaminating eukaryotic RNA.
FIG. 2.
Venn diagrams highlighting disparity between infectious models for genes with altered expression. Numbers indicate the amount of genes determined to have altered expression that are either shared or exclusive to growth of the pneumococci in blood, CSF, or ECC.
Similar articles
- A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae.
McCluskey J, Hinds J, Husain S, Witney A, Mitchell TJ. McCluskey J, et al. Mol Microbiol. 2004 Mar;51(6):1661-75. doi: 10.1111/j.1365-2958.2003.03917.x. Mol Microbiol. 2004. PMID: 15009893 - Site-specific contributions of glutamine-dependent regulator GlnR and GlnR-regulated genes to virulence of Streptococcus pneumoniae.
Hendriksen WT, Kloosterman TG, Bootsma HJ, Estevão S, de Groot R, Kuipers OP, Hermans PW. Hendriksen WT, et al. Infect Immun. 2008 Mar;76(3):1230-8. doi: 10.1128/IAI.01004-07. Epub 2008 Jan 3. Infect Immun. 2008. PMID: 18174343 Free PMC article. - A variable region within the genome of Streptococcus pneumoniae contributes to strain-strain variation in virulence.
Harvey RM, Stroeher UH, Ogunniyi AD, Smith-Vaughan HC, Leach AJ, Paton JC. Harvey RM, et al. PLoS One. 2011 May 5;6(5):e19650. doi: 10.1371/journal.pone.0019650. PLoS One. 2011. PMID: 21573186 Free PMC article. - Role of two-component systems in the virulence of Streptococcus pneumoniae.
Paterson GK, Blue CE, Mitchell TJ. Paterson GK, et al. J Med Microbiol. 2006 Apr;55(Pt 4):355-363. doi: 10.1099/jmm.0.46423-0. J Med Microbiol. 2006. PMID: 16533981 Review. - Strategies for isolation of in vivo expressed genes from bacteria.
Handfield M, Levesque RC. Handfield M, et al. FEMS Microbiol Rev. 1999 Jan;23(1):69-91. doi: 10.1111/j.1574-6976.1999.tb00392.x. FEMS Microbiol Rev. 1999. PMID: 10077854 Review.
Cited by
- IL-27 regulates the differentiation of follicular helper NKT cells via metabolic adaptation of mitochondria.
Kamii Y, Hayashizaki K, Kanno T, Chiba A, Ikegami T, Saito M, Akeda Y, Ohteki T, Kubo M, Yoshida K, Kawakami K, Oishi K, Araya J, Kuwano K, Kronenberg M, Endo Y, Kinjo Y. Kamii Y, et al. Proc Natl Acad Sci U S A. 2024 Feb 27;121(9):e2313964121. doi: 10.1073/pnas.2313964121. Epub 2024 Feb 23. Proc Natl Acad Sci U S A. 2024. PMID: 38394242 Free PMC article. - Exploring metal availability in the natural niche of Streptococcus pneumoniae to discover potential vaccine antigens.
van Beek LF, Surmann K, van den Berg van Saparoea HB, Houben D, Jong WSP, Hentschker C, Ederveen THA, Mitsi E, Ferreira DM, van Opzeeland F, van der Gaast-de Jongh CE, Joosten I, Völker U, Schmidt F, Luirink J, Diavatopoulos DA, de Jonge MI. van Beek LF, et al. Virulence. 2020 Dec;11(1):1310-1328. doi: 10.1080/21505594.2020.1825908. Virulence. 2020. PMID: 33017224 Free PMC article. - Future perspective on host-pathogen interactions during bacterial biofilm formation within the nasopharynx.
Blanchette KA, Orihuela CJ. Blanchette KA, et al. Future Microbiol. 2012 Feb;7(2):227-39. doi: 10.2217/fmb.11.160. Future Microbiol. 2012. PMID: 22324992 Free PMC article. Review. - Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae.
Iyer R, Baliga NS, Camilli A. Iyer R, et al. J Bacteriol. 2005 Dec;187(24):8340-9. doi: 10.1128/JB.187.24.8340-8349.2005. J Bacteriol. 2005. PMID: 16321938 Free PMC article. - Statins protect against fulminant pneumococcal infection and cytolysin toxicity in a mouse model of sickle cell disease.
Rosch JW, Boyd AR, Hinojosa E, Pestina T, Hu Y, Persons DA, Orihuela CJ, Tuomanen EI. Rosch JW, et al. J Clin Invest. 2010 Feb;120(2):627-35. doi: 10.1172/JCI39843. Epub 2010 Jan 19. J Clin Invest. 2010. PMID: 20093777 Free PMC article.
References
- Allen, B. L., and M. Hook. 2002. Isolation of a putative laminin binding protein from Streptococcus anginosus. Microb. Pathog. 33:23-31. - PubMed
- Bethe, G., R. Nau, A. Wellmer, R. Hakenbeck, R. R. Reinert, H. P. Heinz, and G. Zysk. 2001. The cell wall-associated serine protease PrtA: a highly conserved virulence factor of Streptococcus pneumoniae. FEMS Microbiol. Lett. 205:99-104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical