Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex - PubMed (original) (raw)
Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex
Jessica A Sexton et al. Infect Immun. 2004 Oct.
Abstract
Legionella pneumophila utilizes a type IV secretion system (T4SS) encoded by 26 dot/icm genes to replicate inside host cells and cause disease. In contrast to all other L. pneumophila dot/icm genes, dotU and icmF have homologs in a wide variety of gram-negative bacteria, none of which possess a T4SS. Instead, dotU and icmF orthologs are linked to a locus encoding a conserved cluster of proteins designated IcmF-associated homologous proteins, which has been proposed to constitute a novel cell surface structure. We show here that dotU is partially required for L. pneumophila intracellular growth, similar to the known requirement for icmF. In addition, we show that dotU and icmF are necessary for optimal plasmid transfer and sodium sensitivity, two additional phenotypes associated with a functional Dot/Icm complex. We found that these effects are due to the destabilization of the T4SS at the transition into the stationary phase, the point at which L. pneumophila becomes virulent. Specifically, three Dot proteins (DotH, DotG, and DotF) exhibit decreased stability in a DeltadotU DeltaicmF strain. Furthermore, overexpression of just one of these proteins, DotH, is sufficient to suppress the intracellular growth defect of the DeltadotU DeltaicmF mutant. This suggests a model where the DotU and IcmF proteins serve to prevent DotH degradation and therefore function to stabilize the L. pneumophila T4SS. Due to their wide distribution among bacterial species and their genetic linkage to known or predicted cell surface structures, we propose that this function in complex stabilization may be broadly conserved.
Figures
FIG. 1.
IcmF and DotU orthologs are found in IAHP gene clusters. The L. pneumophila (L.p.) dotU and icmF genes (shown by red arrows) are located adjacent to a large number of dot/icm genes (shown by blue arrows) and are immediately flanked by genes with no role in type IV secretion (shown by white arrows). The IAHPs from V. cholerae (V.c.), R. leguminosarum (R.l.), and S. enterica (S.e.) all contain orthologs to dotU and icmF (shown by red arrows). The IAHP clusters contain a conserved core set of proteins (shown by solid green arrows) and less well conserved proteins that are found only in a subset of IAHP loci (indicated by stippled or checkered green arrows). Finally, most of these loci also contain a clpB homolog (shown by a black arrow).
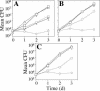
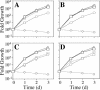
FIG. 2.
Intracellular growth of dotU and icmF mutants in U937 monocytes. (A) The wild-type L. pneumophila strain Lp02 (squares), the dotA mutant Lp03 (diamonds), and the Δ_dotU_ strain JV1116 containing either the empty vector pJB908 (circles), the dotU complementing clone pJB1180 (triangles), or the icmF complementing clone pJB1186 (inverted triangles) were assayed for growth in U937 cells. Mean numbers of CFU are plotted as a function of time. (B) The wild-type L. pneumophila strain Lp02 (squares), the dotA mutant Lp03 (diamonds), and the Δ_icmF_ strain JV1179 containing either the empty vector pJB908 (circles) or the icmF complementing clone pJB1186 (triangles) were assayed for growth in U937 cells. (C) The wild-type L. pneumophila strain Lp02 (squares), the dotA mutant Lp03 (diamonds), and the Δ_dotU_ Δ_icmF_ strain JV1181 containing either the empty vector pJB908 (circles) or the dotU icmF complementing clone pJB1191 (triangles) were assayed for growth in U937 cells. Assays were done in triplicate. Error bars indicate standard deviations of the means. d, day.
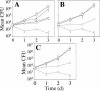
FIG. 3.
Intracellular growth of dotU and icmF mutants in mouse BMM. (A) The wild-type L. pneumophila strain Lp02 (squares), the dotA mutant Lp03 (diamonds), and the Δ_dotU_ strain JV1116 containing either the empty vector pJB908 (circles), the dotU complementing clone pJB1180 (triangles), or the icmF complementing clone pJB1186 (inverted triangles) were assayed for growth in BMM. Mean numbers of CFU are plotted as a function of time. (B) The wild-type L. pneumophila strain Lp02 (squares), the dotA mutant Lp03 (diamonds), and the Δ_icmF_ strain JV1179 containing either the empty vector pJB908 (circles) or the icmF complementing clone pJB1186 (triangles) were assayed for growth in BMM. (C) The wild-type L. pneumophila strain Lp02 (squares), the dotA mutant Lp03 (diamonds), and the Δ_dotU_ Δ_icmF_ strain JV1181 containing either the empty vector pJB908 (circles) or the dotU icmF complementing clone pJB1191 (triangles) were assayed for growth in BMM. Assays were done in triplicate. Error bars indicate standard deviations of the means. d, day.
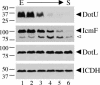
FIG. 4.
DotU and IcmF disappear at different points in stationary-phase L. pneumophila. Equivalent OD600 units of cells from L. pneumophila wild-type strain Lp02 were taken at various time points during growth in broth, from early exponential (E) through late stationary (S) phase. Lanes 1 through 6 correspond to culture OD600s of 2.5, 2.8, 3.1, 3.2, 3.4, and 3.4, respectively. Cells became motile between OD600s of 2.8 and 3.1, just prior to entering the stationary phase. Cell lysates were used for DotU, IcmF, DotL, or ICDH Western blots. Solid arrowheads point to bands that correspond to full-length proteins while the open arrowhead indicates a smaller reactive species likely to represent processed or partially degraded IcmF protein. The molecular masses of relevant markers (in kilodaltons) are shown on the left.
FIG. 5.
Subcellular localization of DotU and IcmF. (A) Total protein (T, lanes 1, 4, and 7), total soluble protein (S, lanes 2, 5, and 8), and total membrane protein (M, lanes 3, 6, and 9) fractions were taken from equivalent OD600 units of three L. pneumophila strains and were subjected to DotU and IcmF Western blots. Strains included the wild-type Lp02 strain (lanes 1 to 3), the Δ_icmF_ strain JV1179 (lanes 4 to 6), and the Δ_dotU_ strain JV4015 (lanes 7 to 9). The molecular masses of relevant markers (in kilodaltons) are shown on the left. Results are representative of those of several experiments.
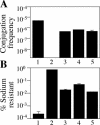
FIG. 6.
Conjugation and salt resistance phenotypes of dotU and icmF mutants. (A) The following L. pneumophila strains were assayed for the ability to transfer an RSF1010 plasmid to E. coli recipient cells: wild-type strain Lp02 (column 1), dotA mutant strain Lp03 (column 2), Δ_dotU_ strain JV1116 (column 3), Δ_icmF_ strain JV1179 (column 4), and Δ_dotU_ Δ_icmF_ strain JV1181 (column 5). The conjugation frequency was calculated as the number of E. coli recipients per L. pneumophila donor cell. Assays were done in triplicate. Error bars indicate the standard deviations of the means. (B) The plating efficiency on 0.65% NaCl was determined for strains used in panel A, and is shown here as the percentage of NaCl-resistant CFU in a cell population. The strains were wild-type Lp02 (column 1), dotA mutant Lp03 (column 2), the Δ_dotU_ strain JV1116 (column 3), the Δ_icmF_ strain JV1179 (column 4), and the Δ_dotU_ Δ_icmF_ strain JV1181 (column 5). Assays were done in triplicate. Error bars indicate standard deviations of the means.
FIG. 7.
DotG, DotF, and DotH protein levels are altered in a Δ_dotU_ Δ_icmF_ strain. Equivalent OD600 units of cells from the L. pneumophila wild-type strain Lp02 (lanes 1 to 4) or the Δ_dotU_ Δ_icmF_ strain JV1181 (lanes 5 to 8) were taken at the early (E), mid-exponential (M), late exponential (L), and stationary (S) phases of growth and used as total protein samples for DotG, DotF, or DotH Western blots. The same samples were used for a DotB Western blot as a negative control. The molecular masses of relevant markers (in kilodaltons) are shown on the left. Solid arrowheads point to bands that correspond to full-length proteins while open arrowheads indicate smaller reactive species likely to represent processed or partially degraded proteins. Results are representative of those of several experiments.
FIG. 8.
Suppression of Δ_dotU_ Δ_icmF_ intracellular growth defect by overexpression of DotH. (A) The wild-type L. pneumophila strain Lp02 (squares), the dotA mutant Lp03 (diamonds), and the Δ_dotU_ Δ_icmF_ strain JV1181 containing either the empty vector pJB908 (circles), the dotU icmF complementing clone pJB1191 (triangles), or the dotHGF complementing clone pJB2132 (hatched squares) were assayed for growth in U937 cells. Mean numbers of CFU are plotted as a function of time. (B) The wild-type L. pneumophila strain Lp02 (squares), the dotA mutant Lp03 (diamonds), and the Δ_dotU_ Δ_icmF_ strain JV1181 containing either the empty vector pJB908 (circles), the dotU icmF complementing clone pJB1191 (triangles), or the dotH complementing clone pJB1555 (hatched squares) were assayed for growth in U937 cells. (C) The wild-type L. pneumophila strain Lp02 (squares), the dotA mutant Lp03 (diamonds), and the Δ_dotU_ Δ_icmF_ strain JV1181 containing either the empty vector pJB908 (circles), the dotU icmF complementing clone pJB1191 (triangles), or the dotG complementing clone pJB1554 (hatched squares) were assayed for growth in U937 cells. (D) The wild-type L. pneumophila strain Lp02 (squares), the dotA mutant Lp03 (diamonds), and the Δ_dotU_ Δ_icmF_ strain JV1181 containing either the empty vector pJB908 (circles), the dotU icmF complementing clone pJB1191 (triangles), or the dotF complementing clone pJB2121 (hatched squares) were assayed for growth in U937 cells. Growth curves were determined in the presence of 100 μM IPTG. Assays were done in triplicate. Error bars indicate standard deviations of the means. d, day.
FIG. 9.
Suppression of Δ_dotU_ Δ_icmF_ protein aberrations by overexpression of DotH. L. pneumophila wild-type strain Lp02 (lanes 1 to 4) or Δ_dotU_ Δ_icmF_ strain JV1181 containing either the empty vector pJB908 (lanes 5 to 8) or the dotH complementing clone pJB1555 (lanes 8 to 12) was grown in the presence of 0, 10, 100, or 1,000 μM IPTG to stationary phase. Whole-cell samples (equivalent OD600 units) were taken and subjected to DotH, DotG, or DotF Western blots. The molecular masses of relevant markers (in kilodaltons) are shown on the left. Arrowheads are as described in the legend to Fig. 7.
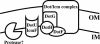
FIG. 10.
Model of how DotU and IcmF prevent destabilization of the Dot/Icm complex. The DotU and IcmF proteins localize to the inner membrane where they likely work together to shield one or more components of the Dot/Icm complex from degradation by an as yet unidentified protease. In the absence of DotU/IcmF, the subcomplex DotH, DotG, and DotF is selectively targeted for proteolysis.
Similar articles
- IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole.
VanRheenen SM, Duménil G, Isberg RR. VanRheenen SM, et al. Infect Immun. 2004 Oct;72(10):5972-82. doi: 10.1128/IAI.72.10.5972-5982.2004. Infect Immun. 2004. PMID: 15385501 Free PMC article. - Characterization of the icmH and icmF genes required for Legionella pneumophila intracellular growth, genes that are present in many bacteria associated with eukaryotic cells.
Zusman T, Feldman M, Halperin E, Segal G. Zusman T, et al. Infect Immun. 2004 Jun;72(6):3398-409. doi: 10.1128/IAI.72.6.3398-3409.2004. Infect Immun. 2004. PMID: 15155646 Free PMC article. - Identification of non-dot/icm suppressors of the Legionella pneumophila DeltadotL lethality phenotype.
Vincent CD, Buscher BA, Friedman JR, Williams LA, Bardill P, Vogel JP. Vincent CD, et al. J Bacteriol. 2006 Dec;188(23):8231-43. doi: 10.1128/JB.00937-06. Epub 2006 Sep 22. J Bacteriol. 2006. PMID: 16997951 Free PMC article. - The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii.
Segal G, Feldman M, Zusman T. Segal G, et al. FEMS Microbiol Rev. 2005 Jan;29(1):65-81. doi: 10.1016/j.femsre.2004.07.001. FEMS Microbiol Rev. 2005. PMID: 15652976 Review. - Effector proteins translocated by Legionella pneumophila: strength in numbers.
Ninio S, Roy CR. Ninio S, et al. Trends Microbiol. 2007 Aug;15(8):372-80. doi: 10.1016/j.tim.2007.06.006. Epub 2007 Jul 13. Trends Microbiol. 2007. PMID: 17632005 Review.
Cited by
- DotU and VgrG, core components of type VI secretion systems, are essential for Francisella LVS pathogenicity.
Bröms JE, Meyer L, Lavander M, Larsson P, Sjöstedt A. Bröms JE, et al. PLoS One. 2012;7(4):e34639. doi: 10.1371/journal.pone.0034639. Epub 2012 Apr 13. PLoS One. 2012. PMID: 22514651 Free PMC article. - Comparative Transcriptomics and Genomics from Continuous Axenic Media Growth Identifies Coxiella burnetii Intracellular Survival Strategies.
Yadav A, Brewer MN, Elshahed MS, Shaw EI. Yadav A, et al. bioRxiv [Preprint]. 2023 Feb 6:2023.02.06.527305. doi: 10.1101/2023.02.06.527305. bioRxiv. 2023. PMID: 36798183 Free PMC article. Updated. Preprint. - Pore formation triggered by Legionella spp. is an Nlrc4 inflammasome-dependent host cell response that precedes pyroptosis.
Silveira TN, Zamboni DS. Silveira TN, et al. Infect Immun. 2010 Mar;78(3):1403-13. doi: 10.1128/IAI.00905-09. Epub 2010 Jan 4. Infect Immun. 2010. PMID: 20048047 Free PMC article. - Advances in genetic manipulation of obligate intracellular bacterial pathogens.
Beare PA, Sandoz KM, Omsland A, Rockey DD, Heinzen RA. Beare PA, et al. Front Microbiol. 2011 May 2;2:97. doi: 10.3389/fmicb.2011.00097. eCollection 2011. Front Microbiol. 2011. PMID: 21833334 Free PMC article. - Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes.
Blondel CJ, Jiménez JC, Contreras I, Santiviago CA. Blondel CJ, et al. BMC Genomics. 2009 Aug 4;10:354. doi: 10.1186/1471-2164-10-354. BMC Genomics. 2009. PMID: 19653904 Free PMC article.
References
- Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. - PubMed
- Bladergroen, M. R., K. Badelt, and H. P. Spaink. 2003. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 16:53-64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI048052/AI/NIAID NIH HHS/United States
- T32 AI007172/AI/NIAID NIH HHS/United States
- 5 T32 AI07172-22/AI/NIAID NIH HHS/United States
- AI48052-02/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources