Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL - PubMed (original) (raw)
Comparative Study
. 2004 Oct 13;23(20):3973-83.
doi: 10.1038/sj.emboj.7600404. Epub 2004 Sep 23.
Affiliations
- PMID: 15385960
- PMCID: PMC524340
- DOI: 10.1038/sj.emboj.7600404
Comparative Study
Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL
Guido Posern et al. EMBO J. 2004.
Abstract
Nuclear accumulation of the serum response factor coactivator MAL/MKL1 is controlled by its interaction with G-actin, which results in its retention in the cytoplasm in cells with low Rho activity. We previously identified actin mutants whose expression promotes MAL nuclear accumulation via an unknown mechanism. Here, we show that actin interacts directly with MAL in vitro with high affinity. We identify a further activating mutation, G15S, which stabilises F-actin, as do the activating actins S14C and V159N. The three mutants share several biochemical properties, but can be distinguished by their ability to bind cofilin, ATP and MAL. MAL interaction with actin S14C is essentially undetectable, and that with actin V159N is weakened. In contrast, actin G15S interacts more strongly with MAL than the wild-type protein. Strikingly, the nuclear accumulation of MAL induced by overexpression of actin S14C is substantially dependent on Rho activity and actin treadmilling, while that induced by actin G15S expression is not. We propose a model in which actin G15S acts directly to promote MAL nuclear entry.
Figures
Figure 1
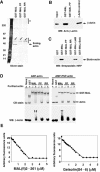
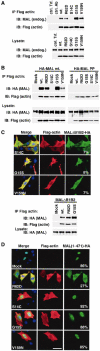
MAL binds directly to actin. (A) Actin does not associate with other cellular proteins on the RPEL motif. NIH3T3 cells were lysed by syringing in detergent-free buffer and the high-speed G-actin supernatant was affinity-precipitated using GST-MAL(met)(1–171) or its derivatives PP34/78AA (PP) or RR33/77DD (RR), which carry point mutations in each of the two RPEL motifs. Bound proteins were eluted with 500 nM swinholide A, separated by 6–16% gradient PAGE and detected by silver staining. The diagonal arrows indicate contaminating GST fusion proteins and asterisks mark nonspecifically retained polypeptides. (B) Precipitated proteins from a GST-MAL(met)(1–171) affinity precipitation experiment of the type shown in (A) were analysed by immunoblotting with β-actin antibody. (C) Affinity precipitation of purified biotinylated nonmuscle actin using GST-MAL(met)(1–171) or its derivatives. Where indicated, 1 μM latrunculin B or 100 nM swinholide A was included in the binding reaction. Bound actin was separated by 12% SDS–PAGE and detected by overlay with peroxidase-conjugated streptavidin. (D) Native 6% polyacrylamide gel electrophoresis assay of complex formation between GST-MAL(met)(1–171) derivatives and ADP- or AMP-PNP-loaded nonmuscle actin (left and right panels, respectively), with detection by Coomassie blue (CB) staining or anti-β-actin immunoblot. (E) Inhibition of skeletal muscle α-actin polymerisation by MAL(fl)2–261 (left), which contains all three RPEL motifs, and gelsolin(S4–6) (right). Data are mean of two independent experiments.
Figure 2
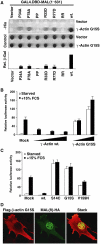
Identification of actin G15S, an activator of MAL and SRF. (A) Yeast two-hybrid interactions between γ-actin G15S and MAL(met)(1–631) or mutant derivatives containing point changes in either or both RPEL motifs. Upper panels: _trans_-illuminated images of colony growth on selective (−His) or nonselective (control) medium. Lower histogram: interaction quantification by liquid-culture Gal4-lacZ reporter gene assay (WT γ-actin=100; error bars: s.e.m.; _n_=3) (B) γ-Actin G15S expression activates SRF. Cells were transfected with SRF reporter 3D.A-Luc (40 ng), together with wild-type γ-actin (100, 250 or 500 ng) or γ-actin G15S (100 or 250 ng), maintained in 0.3% FCS for 40 h and then serum-stimulated where indicated. Data are means of three independent experiments; error bars: s.e.m. (C) Reporter activation by β-actin G15S and other β-actin mutants. Cells were transfected with reporter and the indicated β-actin mutants and treated as in (B). (D) β-Actin G15S expression induces MAL nuclear accumulation. Cells expressing MAL(fl)-HA (50 ng) and β-actin G15S (500 ng) were processed for immunofluorescence 24 h after transfection. Confocal sections of 0.3 μm thickness show actin G15S (anti-Flag; red) and nuclear accumulation of MAL(fl) (anti-HA; green). The merged picture shows the stack of both actin and MAL sections.
Figure 3
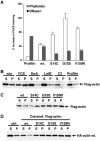
Actin G15S stabilises filament formation. (A) Transfected cells expressing the indicated Flag-tagged actins (1 μg) were stained for the Flag epitope and either TRITC-phalloidin (for F-actin) or FITC-DNase I (for G-actin). The FACS was used to quantify mean levels of F- or G-actin in the transfected population relative to those of the untransfected population (error bars: s.e.m.; _n_=3). (B) Actin fractionation lysates were prepared from cells expressing wild-type Flag-actin (1 μg) with C3 transferase (50 ng) or profilin (500 ng) coexpression, or treatment with latrunculin B (0.3 μM, 1 h), swinholide A (100 nM, 1 h) or FCS (15%, 10 min) as indicated. Flag-actin in each supernatant (S) and pellet (P) fraction was detected by immunoblotting using anti-Flag antibodies. (C) Actin fractionation lysates were prepared from cells expressing the indicated Flag-tagged actin mutants (1 μg) and analysed by anti-Flag immunoblotting as in (B). (D) Activating actin mutants copolymerise with and stabilise wild-type F-actin. Actin fractionation lysates were prepared from cells expressing the indicated Flag-tagged actins (1 μg) together with HA-tagged wild-type actin (500 ng), and analysed by anti-HA immunoblot.
Figure 4
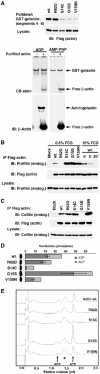
Activating actin mutants have both shared and distinct properties. (A) Activating actins exhibit reduced affinity for the C-terminal half of gelsolin. Upper panel: G-actin supernatants from cells expressing the indicated actins (2 μg) were affinity-precipitated using GST-gelsolin(S4–6), and bound proteins were detected by immunoblotting with anti-Flag antibodies. Lower panels: native gel electrophoresis assay of complex formation between GST-gelsolin(S4–6) and ADP- or AMP-PNP-loaded nonmuscle actin, with detection by Coomassie blue or anti-β-actin immunoblot. (B) Interaction of actins with endogenous profilin. Upper panels: Extracts as in (A) were immunoprecipitated with anti-Flag and analysed for profilin (anti-profilin; upper) and actin (anti-Flag; lower). Cells were in 0.5% serum except where stimulated with 15% serum for the indicated times (min). Lower panel: Control immunoblot for profilin in the lysate. (C) Interaction of actins with endogenous cofilin. Extracts were prepared and analysed as in (B). (D) Nucleotide binding by actin mutants. Actin immunoprecipitates were prepared as in (B), followed by nucleotide determination using the luciferase-based ATP assay. Immunoblot: recovered actin in each sample. Data are mean±s.e.m. (_n_=4). (E) Nucleotide binding by actin mutants. Actins were prepared as in (D) and nucleotide was identified by Mono-Q anion exchange chromatography. UV absorbance at 254 nm from a representative experiment is shown, with elution volumes for ADP and ATP indicated. The asterisk indicates a nonspecifically recovered component also present in precipitates from mock-transfected cells.
Figure 5
Interaction of MAL with wild-type and mutant actins. (A) Interaction of actins with endogenous MAL from NIH3T3 cells. Upper panels: extracts from cells expressing the indicated Flag-tagged actin mutants (1 μg) were immunoprecipitated with anti-Flag or control antibodies, and analysed for MAL (anti-MAL; upper) and actin (anti-Flag; lower). Lower panels: control immunoblots for MAL (anti-MAL; upper) and actin expression (anti-Flag) in the lysate. ctrl. Trf.: no actin; ctrl. Ab: wild-type actin, anti-HA IP. (B) Interaction of actins with overexpressed HA-tagged MAL(met) and HA-MAL(met) PP34/78AA (containing mutated RPEL motifs) (1 μg). Cells were transfected and processed as in (A). Data in lanes 1–3 are from Miralles et al (2003). (C) Nuclear accumulation induced by the activating actins requires MAL basic box regions. Upper panels: cells in 0.5% FCS expressing MALΔB1B2-HA (0.1 μg), which lacks the basic regions, and activating actins (1 μg) were fixed and stained for MAL (anti-HA; green), actins (anti-Flag; red) and DNA (Hoechst 33258; blue). Numbers show proportion of cells with predominantly nuclear MAL. Lower panels: control co-immunoprecipitation experiments performed as in (B). (D) Overexpression of nonpolymerisable actin R62D, but not the F-actin-stabilising mutants, promotes relocalisation of MAL(met)1–471 to the cytoplasm. Cells expressing HA-tagged MAL(met)1–471 with vector (mock) or Flag-tagged actins were stained and scored as in (C).
Figure 6
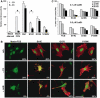
Activating actins exhibit different requirements for actin treadmilling in SRF activation. (A) SRF reporter activation by activating actins is differentially sensitive to Rho inactivation. Cells transfected with SRF reporter expressed activating actins (500 ng), C3 transferase (20 ng) and activated mDia (50 ng) as indicated. Data are the mean of four independent experiments (error bars: s.e.m.; asterisks: statistical significance at P<0.01, unpaired Student's _t_-test). (B) Immunofluorescence analysis of MAL nuclear accumulation induced by serum stimulation or coexpression with activating actin mutants. Cells expressed MAL(met) (50 ng) and activating actins (500 ng), with C3 transferase coexpression (20 ng) or latrunculin B treatment as indicated, and were maintained in 0.5% FCS unless stimulated with 15% FCS for 30 min as indicated. Cells were fixed and stained for MAL (anti-HA; green) and actins (anti-Flag; red). Latrunculin treatment (0.3 μM) was for 30 min prior to staining. Around 100 cells each were analysed and scored for predominantly nuclear MAL staining. (C) Kinetics of MAL cytoplasmic reaccumulation. Cells were transfected as in (B) and treated with either 0.1 or 0.3 μM latrunculin B for the times indicated. Data are the mean percentage of predominantly nuclear MAL staining in two independent experiments.
Figure 7
(A) Summary of mutant actin properties. Biochemical characteristics of the indicated point mutants are compared to those of the wild-type protein. (B) Models for activation of MAL by mutant actins. (i) Normal cells in low serum exhibit a basal actin treadmilling (circle; G: G-actin; F: F-actin). MAL is retained in the cytoplasm (open box) by interaction with G-actin (white circles), which somehow inhibits its nuclear accumulation. Upon serum stimulation, the G-actin pool is depleted and MAL released for nuclear import (black box); blockade of treadmilling by latrunculin B or Rho inactivation prevents this, inhibiting MAL release (open box). (ii) Actin S14C (black circles) stabilises F-actin; its overexpression increases F-actin but does not deplete the G-actin pool. However, actin S14C itself does not effectively bind MAL, which is therefore released for nuclear accumulation (black box). Upon blockade of treadmilling, wild-type G-actin reaccumulates, and MAL is again inhibited (open box). (iii) Actin G15S (black triangles) stabilises F-actin; its overexpression increases F-actin but does not deplete the G-actin pool. Binding of actin G15S to MAL directly promotes MAL nuclear accumulation (black box). MAL therefore remains active upon inhibition of actin treadmilling.
Similar articles
- Actin dynamics control SRF activity by regulation of its coactivator MAL.
Miralles F, Posern G, Zaromytidou AI, Treisman R. Miralles F, et al. Cell. 2003 May 2;113(3):329-42. doi: 10.1016/s0092-8674(03)00278-2. Cell. 2003. PMID: 12732141 - RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding.
Guettler S, Vartiainen MK, Miralles F, Larijani B, Treisman R. Guettler S, et al. Mol Cell Biol. 2008 Jan;28(2):732-42. doi: 10.1128/MCB.01623-07. Epub 2007 Nov 19. Mol Cell Biol. 2008. PMID: 18025109 Free PMC article. - Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL.
Mouilleron S, Guettler S, Langer CA, Treisman R, McDonald NQ. Mouilleron S, et al. EMBO J. 2008 Dec 3;27(23):3198-208. doi: 10.1038/emboj.2008.235. Epub 2008 Nov 13. EMBO J. 2008. PMID: 19008859 Free PMC article. - A nuclear MAL-function links Rho to SRF.
Settleman J. Settleman J. Mol Cell. 2003 May;11(5):1121-3. doi: 10.1016/s1097-2765(03)00189-8. Mol Cell. 2003. PMID: 12769835 Review. - Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression.
Cen B, Selvaraj A, Prywes R. Cen B, et al. J Cell Biochem. 2004 Sep 1;93(1):74-82. doi: 10.1002/jcb.20199. J Cell Biochem. 2004. PMID: 15352164 Review.
Cited by
- Single-molecule tracking (SMT) and localization of SRF and MRTF transcription factors during neuronal stimulation and differentiation.
Kuchler O, Gerlach J, Vomhof T, Hettich J, Steinmetz J, Gebhardt JCM, Michaelis J, Knöll B. Kuchler O, et al. Open Biol. 2022 May;12(5):210383. doi: 10.1098/rsob.210383. Epub 2022 May 11. Open Biol. 2022. PMID: 35537478 Free PMC article. - Loss of serum response factor in mature neurons in the dentate gyrus alters the morphology of dendritic spines and hippocampus-dependent behavioral tasks.
Nader K, Krysiak A, Beroun A, Pekala M, Szymanska M, Kuzniewska B, Radwanska K, Kaczmarek L, Kalita K. Nader K, et al. Brain Struct Funct. 2019 Nov;224(8):2691-2701. doi: 10.1007/s00429-019-01925-6. Epub 2019 Aug 2. Brain Struct Funct. 2019. PMID: 31375980 Free PMC article. - SRF'ing and SAP'ing - the role of MRTF proteins in cell migration.
Gau D, Roy P. Gau D, et al. J Cell Sci. 2018 Oct 11;131(19):jcs218222. doi: 10.1242/jcs.218222. J Cell Sci. 2018. PMID: 30309957 Free PMC article. Review. - Myocardin-Related Transcription Factor A Activation by Competition with WH2 Domain Proteins for Actin Binding.
Weissbach J, Schikora F, Weber A, Kessels M, Posern G. Weissbach J, et al. Mol Cell Biol. 2016 May 2;36(10):1526-39. doi: 10.1128/MCB.01097-15. Print 2016 May 15. Mol Cell Biol. 2016. PMID: 26976641 Free PMC article. - Structure-function analysis of the role of megakaryoblastic leukemia 1 in megakaryocyte polyploidization.
Reed FE, Eskow NM, Min E, Carlino M, Mancuso R, Kwon N, Smith EC, Larsuel ST, Wang L, Scanlon V, Krause DS. Reed FE, et al. Haematologica. 2022 Dec 1;107(12):2972-2976. doi: 10.3324/haematol.2021.280499. Haematologica. 2022. PMID: 36453520 Free PMC article. No abstract available.
References
- Belmont LD, Patterson GM, Drubin DG (1999b) New actin mutants allow further characterization of the nucleotide binding cleft and drug binding sites. J Cell Sci 112: 1325–1336 - PubMed
- Bettinger BT, Gilbert DM, Amberg DC (2004) Actin up in the nucleus. Nat Rev Mol Cell Biol 5: 410–415 - PubMed
- Bourne HR, Sanders DA, McCormick F (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349: 117–127 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous