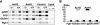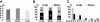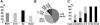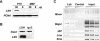Lsh, an epigenetic guardian of repetitive elements - PubMed (original) (raw)
. 2004 Sep 24;32(17):5019-28.
doi: 10.1093/nar/gkh821. Print 2004.
Affiliations
- PMID: 15448183
- PMCID: PMC521642
- DOI: 10.1093/nar/gkh821
Lsh, an epigenetic guardian of repetitive elements
Jiaqiang Huang et al. Nucleic Acids Res. 2004.
Abstract
The genome is burdened with repetitive sequences that are generally embedded in silenced chromatin. We have previously demonstrated that Lsh (lymphoid-specific helicase) is crucial for the control of heterochromatin at pericentromeric regions consisting of satellite repeats. In this study, we searched for additional genomic targets of Lsh by examining the effects of Lsh deletion on repeat regions and single copy gene sequences. We found that the absence of Lsh resulted in an increased association of acetylated histones with repeat sequences and transcriptional reactivation of their silenced state. In contrast, selected single copy genes displayed no change in histone acetylation levels, and their transcriptional rate was indistinguishable compared to Lsh-deficient cells and wild-type controls. Microarray analysis of total RNA derived from brain and liver tissues revealed that <0.4% of the 15 247 examined loci were abnormally expressed in Lsh-/-embryos and almost two-thirds of these deregulated sequences contained repeats, mainly retroviral LTR (long terminal repeat) elements. Chromatin immunoprecipitation analysis demonstrated a direct interaction of Lsh with repetitive sites in the genome. These data suggest that the repetitive sites are direct targets of Lsh action and that Lsh plays an important role as 'epigenetic guardian' of the genome to protect against deregulation of parasitic retroviral elements.
Figures
Figure 1
Increased association of acetylated histones with repetitive elements in Lsh-deficient cells. (A) ChIP assay was performed from Lsh-deficient and wild-type chromatin extracts using indicated antibodies (normal rabbit IgG as control) to detect acetylation of histone 3 (Lys-9/14) or histone 4 (Lys-5/8/12/16). Precipitated DNA or chromatin input was diluted (1:3) for the detection of linear PCR amplification followed by Southern analysis. (B) Results of ChIPs were quantified using a PhosporImager, and the ratio of Lsh−/− to wild-type controls was calculated. The average values of four to five experiments are plotted in the bar graph.
Figure 2
Unchanged histone acetylation levels at single copy sequences. ChIP assay was performed using antibodies as indicated to detect acetylation of histone 3 (Lys-9/14) or histone 4 (Lys-5/8/12/16) in nuclear extracts derived from Lsh−/−, Lsh+/+ MEFs followed by PCR analysis. (A) Southern analysis. (B) Taqman-PCR analysis. The average values were calculated from four experiments.
Figure 3
Deregulation of transcripts with repetitive elements in the absence of Lsh. (A) Total RNA derived from Lsh−/− or Lsh+/+ MEF cells or Lsh−/− and Lsh+/+ embryos of day 13.5 of gestation was subjected to RT–PCR analysis. Repeat sequences were analyzed in dilutions of 1:3, single copy genes in dilutions of 1:10. Omission of reverse transcriptase (−RT) or treatment with RNase served as further controls. (B) Analysis of strand-specific transcripts using forward and reverse primers during reverse transcription. Oligo(dt) primed cDNA served as control for detection of β-actin and IAP transcripts.
Figure 4
Microarray analysis of gene expression after Lsh deletion. Total RNA derived from Lsh−/− or Lsh+/+ liver or brain tissue of day 17.5 gestation embryos were subjected to Microarray analysis. Results represent the average of three hybridizations performed for each tissue type. (A) Proportion of clones from the whole microarray that are abnormally up- or downregulated more than 2-fold above wild-type controls. (B) Proportion of deregulated genes that are either up- or downregulated in the brain or liver tissues. (C) Proportion of deregulated genes that are up- or downregulated and their degree of deregulation expressed as a fold above wild-type control.
Figure 5
Presence of repeat elements in deregulated sequences. (A) Proportion of deregulated clones in brain or liver tissues that contain a repeat element. (B) Proportion of clones that contain different types of repeat elements. Note that a few clones contain more than one type of repeat elements. About 9% of clones did contain neither LTR elements, SINE, LINE nor DNA elements, and only simple repeats were present. (C) Whole bars indicate the proportion of deregulated sequences that contain repeats in relation to their magnitude of deregulation. Black bars indicate the proportion of deregulated sequences containing an LTR.
Figure 6
Lsh targets to repetitive sequences. (A) Western analysis of Lsh expression in the embryonal carcinoma cell-line P19 and embryonal fibroblasts (MEFs) by using a specific antibody raised against the C-terminal portion of Lsh. (B) ChIPs were performed on P19 cells using the C-terminal antibody against Lsh or an IgG control. The precipitated material was examined by western analysis using the C-terminal anti-Lsh antibody for detection. (C) ChIPs were performed from P19 chromatin extracts using the C-terminal antibody against Lsh or an IgG control. Precipitated DNA or chromatin input was analyzed for the presence of the indicated repeat sequences. DNA was diluted (1:3) for the detection of linear PCR amplification.
Similar articles
- Lsh, a guardian of heterochromatin at repeat elements.
Muegge K. Muegge K. Biochem Cell Biol. 2005 Aug;83(4):548-54. doi: 10.1139/o05-119. Biochem Cell Biol. 2005. PMID: 16094458 Review. - Lsh regulates LTR retrotransposon repression independently of Dnmt3b function.
Dunican DS, Cruickshanks HA, Suzuki M, Semple CA, Davey T, Arceci RJ, Greally J, Adams IR, Meehan RR. Dunican DS, et al. Genome Biol. 2013 Dec 24;14(12):R146. doi: 10.1186/gb-2013-14-12-r146. Genome Biol. 2013. PMID: 24367978 Free PMC article. - Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells.
De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. De La Fuente R, et al. Nat Cell Biol. 2006 Dec;8(12):1448-54. doi: 10.1038/ncb1513. Epub 2006 Nov 19. Nat Cell Biol. 2006. PMID: 17115026 - Genome-wide DNA methylation patterns in LSH mutant reveals de-repression of repeat elements and redundant epigenetic silencing pathways.
Yu W, McIntosh C, Lister R, Zhu I, Han Y, Ren J, Landsman D, Lee E, Briones V, Terashima M, Leighty R, Ecker JR, Muegge K. Yu W, et al. Genome Res. 2014 Oct;24(10):1613-23. doi: 10.1101/gr.172015.114. Epub 2014 Aug 28. Genome Res. 2014. PMID: 25170028 Free PMC article. - The ghosts in the machine: DNA methylation and the mystery of differentiation.
Briones V, Muegge K. Briones V, et al. Biochim Biophys Acta. 2012 Jul;1819(7):757-62. doi: 10.1016/j.bbagrm.2012.02.013. Epub 2012 Feb 22. Biochim Biophys Acta. 2012. PMID: 22381140 Free PMC article. Review.
Cited by
- DNA CpG methylation in sequential glioblastoma specimens.
Kraboth Z, Galik B, Tompa M, Kajtar B, Urban P, Gyenesei A, Miseta A, Kalman B. Kraboth Z, et al. J Cancer Res Clin Oncol. 2020 Nov;146(11):2885-2896. doi: 10.1007/s00432-020-03349-w. Epub 2020 Aug 10. J Cancer Res Clin Oncol. 2020. PMID: 32779022 Free PMC article. - Chromatin Remodeling Factor LSH is Upregulated by the LRP6-GSK3β-E2F1 Axis Linking Reversely with Survival in Gliomas.
Xiao D, Huang J, Pan Y, Li H, Fu C, Mao C, Cheng Y, Shi Y, Chen L, Jiang Y, Yang R, Liu Y, Zhou J, Cao Y, Liu S, Tao Y. Xiao D, et al. Theranostics. 2017 Jan 1;7(1):132-143. doi: 10.7150/thno.17032. eCollection 2017. Theranostics. 2017. PMID: 28042322 Free PMC article. - Lsh mediated RNA polymerase II stalling at HoxC6 and HoxC8 involves DNA methylation.
Tao Y, Xi S, Briones V, Muegge K. Tao Y, et al. PLoS One. 2010 Feb 11;5(2):e9163. doi: 10.1371/journal.pone.0009163. PLoS One. 2010. PMID: 20161795 Free PMC article. - Reduced syncytin-1 expression levels in placental syndromes correlates with epigenetic hypermethylation of the ERVW-1 promoter region.
Ruebner M, Strissel PL, Ekici AB, Stiegler E, Dammer U, Goecke TW, Faschingbauer F, Fahlbusch FB, Beckmann MW, Strick R. Ruebner M, et al. PLoS One. 2013;8(2):e56145. doi: 10.1371/journal.pone.0056145. Epub 2013 Feb 14. PLoS One. 2013. PMID: 23457515 Free PMC article. - Constitutive heterochromatin formation and transcription in mammals.
Saksouk N, Simboeck E, Déjardin J. Saksouk N, et al. Epigenetics Chromatin. 2015 Jan 15;8:3. doi: 10.1186/1756-8935-8-3. eCollection 2015. Epigenetics Chromatin. 2015. PMID: 25788984 Free PMC article. Review.
References
- Felsenfeld G. and Groudine,M. (2003) Controlling the double helix. Nature, 421, 448–453. - PubMed
- Khorasanizadeh S. (2004) The nucleosome: from genomic organization to genomic regulation. Cell, 116, 259–272. - PubMed
- Fahrner J.A. and Baylin,S.B. (2003) Heterochromatin: stable and unstable invasions at home and abroad. Genes Dev., 17, 1805–1812. - PubMed
- Henikoff S. (2000) Heterochromatin function in complex genomes. Biochim. Biophys. Acta, 1470, O1–O8. - PubMed
- Hashimshony T., Zhang,J., Keshet,I., Bustin,M. and Cedar,H. (2003) The role of DNA methylation in setting up chromatin structure during development. Nature Genet., 34, 187–192. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases