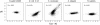Solving the riddle of codon usage preferences: a test for translational selection - PubMed (original) (raw)
. 2004 Sep 24;32(17):5036-44.
doi: 10.1093/nar/gkh834. Print 2004.
Affiliations
- PMID: 15448185
- PMCID: PMC521650
- DOI: 10.1093/nar/gkh834
Solving the riddle of codon usage preferences: a test for translational selection
Mario dos Reis et al. Nucleic Acids Res. 2004.
Abstract
Translational selection is responsible for the unequal usage of synonymous codons in protein coding genes in a wide variety of organisms. It is one of the most subtle and pervasive forces of molecular evolution, yet, establishing the underlying causes for its idiosyncratic behaviour across living kingdoms has proven elusive to researchers over the past 20 years. In this study, a statistical model for measuring translational selection in any given genome is developed, and the test is applied to 126 fully sequenced genomes, ranging from archaea to eukaryotes. It is shown that tRNA gene redundancy and genome size are interacting forces that ultimately determine the action of translational selection, and that an optimal genome size exists for which this kind of selection is maximal. Accordingly, genome size also presents upper and lower boundaries beyond which selection on codon usage is not possible. We propose a model where the coevolution of genome size and tRNA genes explains the observed patterns in translational selection in all living organisms. This model finally unifies our understanding of codon usage across prokaryotes and eukaryotes. Helicobacter pylori, Saccharomyces cerevisiae and Homo sapiens are codon usage paradigms that can be better understood under the proposed model.
Figures
Figure 1
Genetic code and general codon–anticodon recognition rules for tRNA genes. This table simply summarizes all the theoretically possible interactions between the coding codons and the extant tRNA sequences in the organisms analysed in this work. The interested reader is advised to refer to the literature (47,46) for a detailed description of codon–anticodon pairings.
Figure 2
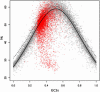
Nc-plot for yeast and simulated E.coli K12 genes. Grey points, simulated E.coli K12 genes; red points, actual yeast genes; dashed line, Wright's proposed function (Equation 5); bold line, the function proposed herein (Equation 8) with optimized parameters.
Figure 3
tAI versus _f_1(x)–Nc for five organisms for which codon usage has been well studied.
Figure 4
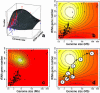
Action of natural selection on codon usage in the genomic landscape. (a) Fitted regression surface of _S_-values to tRNA gene number and genome size. Pink dots, predicted _S_-values for every organism; red dots, organisms with _S_-values higher than predicted by the model; blue, organisms with S-values lower than predicted. Vertical lines join each observed data point to its predicted value. (b) Thermal image (contour plot) of the same regression surface. The hottest (highest) _S_-values are shown in white, while the cooler (lowest) values are in red. E, Eukaryota; B, Eubacteria; and A, Archaea. The contour lines reflect the estimated _S_-value for a particular region. (c) Estimated probability density function for the presence of a maximum in the regression analysis. (d) Hypothetical evolution of codon usage optimization. A small genome sized ancestor (0), suffered a series of genome expansions (1–4). During this evolutionary process, the phylogeny would move into, and then out of the selective hot-spot. The process can be reverted at any time if selection for genome size or tRNA set reduction is present, such as in non-free living organisms e.g. H.pylori (5) or P.falciparum (6).
Similar articles
- Estimating translational selection in eukaryotic genomes.
dos Reis M, Wernisch L. dos Reis M, et al. Mol Biol Evol. 2009 Feb;26(2):451-61. doi: 10.1093/molbev/msn272. Epub 2008 Nov 25. Mol Biol Evol. 2009. PMID: 19033257 Free PMC article. - Synonymous codon ordering: a subtle but prevalent strategy of bacteria to improve translational efficiency.
Shao ZQ, Zhang YM, Feng XY, Wang B, Chen JQ. Shao ZQ, et al. PLoS One. 2012;7(3):e33547. doi: 10.1371/journal.pone.0033547. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22432034 Free PMC article. - Codon usage bias.
Parvathy ST, Udayasuriyan V, Bhadana V. Parvathy ST, et al. Mol Biol Rep. 2022 Jan;49(1):539-565. doi: 10.1007/s11033-021-06749-4. Epub 2021 Nov 25. Mol Biol Rep. 2022. PMID: 34822069 Free PMC article. Review. - The Yin and Yang of codon usage.
Komar AA. Komar AA. Hum Mol Genet. 2016 Oct 1;25(R2):R77-R85. doi: 10.1093/hmg/ddw207. Epub 2016 Jun 27. Hum Mol Genet. 2016. PMID: 27354349 Free PMC article. Review.
Cited by
- Link Between Individual Codon Frequencies and Protein Expression: Going Beyond Codon Adaptation Index.
Zaytsev K, Bogatyreva N, Fedorov A. Zaytsev K, et al. Int J Mol Sci. 2024 Oct 29;25(21):11622. doi: 10.3390/ijms252111622. Int J Mol Sci. 2024. PMID: 39519173 Free PMC article. - SARS-CoV-2 Displays a Suboptimal Codon Usage Bias for Efficient Translation in Human Cells Diverted by Hijacking the tRNA Epitranscriptome.
Eldin P, David A, Hirtz C, Battini JL, Briant L. Eldin P, et al. Int J Mol Sci. 2024 Oct 29;25(21):11614. doi: 10.3390/ijms252111614. Int J Mol Sci. 2024. PMID: 39519170 Free PMC article. - Host-dependent C-to-U RNA editing in SARS-CoV-2 creates novel viral genes with optimized expressibility.
Zhang P, Zhang W, Li J, Liu H, Yu Y, Yang X, Jiang W. Zhang P, et al. Front Cell Infect Microbiol. 2024 Oct 9;14:1476605. doi: 10.3389/fcimb.2024.1476605. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39445213 Free PMC article. - A machine learning approach uncovers principles and determinants of eukaryotic ribosome pausing.
Aguilar Rangel M, Stein K, Frydman J. Aguilar Rangel M, et al. Sci Adv. 2024 Oct 18;10(42):eado0738. doi: 10.1126/sciadv.ado0738. Epub 2024 Oct 18. Sci Adv. 2024. PMID: 39423268 Free PMC article. - The dynamics and functional impact of tRNA repertoires during early embryogenesis in zebrafish.
Reimão-Pinto MM, Behrens A, Forcelloni S, Fröhlich K, Kaya S, Nedialkova DD. Reimão-Pinto MM, et al. EMBO J. 2024 Oct 14. doi: 10.1038/s44318-024-00265-4. Online ahead of print. EMBO J. 2024. PMID: 39402326
References
- Kimura M. (1968) Evolutionary rate at the molecular level. Nature, 217, 624–626. - PubMed
- King J.L. and Jukes,T.H. (1969) Non-Darwinian evolution. Science, 164, 788–798. - PubMed
- Clarke B. (1970) Darwinian evolution of proteins. Science, 168, 1009–1011. - PubMed
- Post L.E. and Nomura,M. (1980) DNA sequences from the str operon of Escherichia coli. J. Biol. Chem., 255, 4660–4665. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous