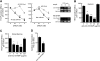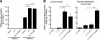RANKL-induced DC-STAMP is essential for osteoclastogenesis - PubMed (original) (raw)
Comparative Study
. 2004 Oct 4;200(7):941-6.
doi: 10.1084/jem.20040518. Epub 2004 Sep 27.
Naohisa Wada, Akiko Kukita, Takashi Kakimoto, Ferry Sandra, Kazuko Toh, Kengo Nagata, Tadahiko Iijima, Madoka Horiuchi, Hiromi Matsusaki, Kunio Hieshima, Osamu Yoshie, Hisayuki Nomiyama
Affiliations
- PMID: 15452179
- PMCID: PMC2213286
- DOI: 10.1084/jem.20040518
Comparative Study
RANKL-induced DC-STAMP is essential for osteoclastogenesis
Toshio Kukita et al. J Exp Med. 2004.
Abstract
Osteoclasts are bone-resorbing, multinucleated giant cells that are essential for bone remodeling and are formed through cell fusion of mononuclear precursor cells. Although receptor activator of nuclear factor-kappaB ligand (RANKL) has been demonstrated to be an important osteoclastogenic cytokine, the cell surface molecules involved in osteoclastogenesis are mostly unknown. Here, we report that the seven-transmembrane receptor-like molecule, dendritic cell-specific transmembrane protein (DC-STAMP) is involved in osteoclastogenesis. Expression of DC-STAMP is rapidly induced in osteoclast precursor cells by RANKL and other osteoclastogenic stimulations. Targeted inhibition of DC-STAMP by small interfering RNAs and specific antibody markedly suppressed the formation of multinucleated osteoclast-like cells. Overexpression of DC-STAMP enhanced osteoclastogenesis in the presence of RANKL. Furthermore, DC-STAMP directly induced the expression of the osteoclast marker tartrate-resistant acid phosphatase. These data demonstrate for the first time that DC-STAMP has an essential role in osteoclastogenesis.
Figures
Figure 1.
Induction of DC-STAMP and its splicing variant DC-STAMPΔT7 in osteoclastogenesis. (A) Structures of DC-STAMP and DC-STAMPΔT7. Noncoding and coding sequences are shown in unshaded and shaded boxes, respectively. PCR primers used in C and D are indicated by arrows. Schematic protein structures are shown below the gene. Shaded regions indicate the putative transmembrane domains. (B) Northern blot analysis for induction of DC-STAMP and DC-STAMPΔT7 in RAW-D cells. RAW-D and RAW-N cells were stimulated for 72 h as indicated. c, control; N, RAW-N; D, RAW-D. (C) RT-PCR analysis on the time course of DC-STAMP and DC-STAMPΔT7 induction in RAW-D cells. RAW-D cells were stimulated with RANKL + TNF-α for indicated periods of time. Expression of DC-STAMP, DC-STAMPΔT7, and osteoclast marker genes (cathepsin K and TRAP) was analyzed. (D) RT-PCR analysis on the time course of DC-STAMP and DC-STAMPΔT7 induction in mouse BM cells. Mouse BM cells were stimulated with 1α,25(OH)2D3. Expression of DC-STAMP and DC-STAMPΔT7 was analyzed. The GenBank/EMBL/DDBJ accession nos. of mouse DC-STAMP and DC-STAMPΔT7 are AB109560 and AB109561, respectively.
Figure 2.
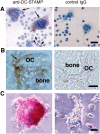
Immunological staining of DC-STAMP in osteoclasts. Cells and tissues were stained with anti–DC-STAMP (left) or control preimmune IgG (right). (A) RAW-D cells stimulated with RANKL + TNF-α for 3 d. Nuclei were visualized by staining with hematoxylin. DC-STAMP positive osteoclast-like MNCs and mononuclear cells are indicated by arrows and arrowheads, respectively. (B) Osteoclasts present in the mandibular tissue of newborn mice. (C) Osteoclasts isolated from the tibia of newborn mice. OC, osteoclast. Bars: 25 μm (A), 10 μm (B), and 20 μm (C).
Figure 3.
Inhibition of osteoclastogenesis by DC-STAMP siRNAs and by anti–DC-STAMP. (A) Effects of the siRNAs on the formation of osteoclast-like TRAP positive MNCs in RAW-D cells stimulated with RANKL and TNF-α for 3 d. Specific reduction of DC-STAMP mRNA and DC-STAMPΔT7mRNA by #6 siRNA was evaluated by RT-PCR (right). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Inhibition of osteoclast-like TRAP positive MNC formation in RAW-D cells by anti–DC-STAMP. RAW-D cells were treated with RANKL and TNF-α for 3 d without or with indicated concentrations of anti–DC-STAMP. **, P < 0.01; ***, P < 0.001. IgG, control IgG (20 μg/ml). (C) Inhibition of osteoclastogenesis in mouse BM cells by anti–DC-STAMP. BM cells were cultured in the presence of 1α,25(OH)2D3 for 6 d without or with indicated concentrations of anti–DC-STAMP. *, P < 0.05; **, P < 0.01. (D) Anti–DC-STAMP inhibits osteoclast function. Rat osteoclast-like cells formed in BM cultures were seeded on dentin slices and cultured for 3 d in the presence of 10 μg/ml of control IgG or anti–DC-STAMP. *, P < 0.05.
Figure 4.
Promotion of osteoclastogenesis in RAW-D cells by DC-STAMP and DC-STAMPΔT7. (A) Transfection of RAW-D cells with control, DC-STAMP, or DC-STAMPΔT7 expression vectors. Cells were treated without or with RANKL + TNF-α for 3 d. Osteoclast-like TRAP positive MNCs were counted. **, P < 0.01; ***, P < 0.001. (B) Induction of TRAP in RAW-D cells by coculture with mouse pre–B L1.2 cells stably expressing DC-STAMP or DC-STAMPΔT7. RAW-D cells were cocultured with live (left) or fixed (right) L1.2 cells or L1.2 cells stably expressing DC-STAMP or DC-STAMPΔT7 in the absence of osteoclastogenic factors for 3 d. Osteoclast-like TRAP positive MNCs were counted. **, P < 0.01.
Similar articles
- RANKL induces heterogeneous DC-STAMP(lo) and DC-STAMP(hi) osteoclast precursors of which the DC-STAMP(lo) precursors are the master fusogens.
Mensah KA, Ritchlin CT, Schwarz EM. Mensah KA, et al. J Cell Physiol. 2010 Apr;223(1):76-83. doi: 10.1002/jcp.22012. J Cell Physiol. 2010. PMID: 20039274 Free PMC article. - Osteoclast stimulatory transmembrane protein (OC-STAMP), a novel protein induced by RANKL that promotes osteoclast differentiation.
Yang M, Birnbaum MJ, MacKay CA, Mason-Savas A, Thompson B, Odgren PR. Yang M, et al. J Cell Physiol. 2008 May;215(2):497-505. doi: 10.1002/jcp.21331. J Cell Physiol. 2008. PMID: 18064667 Free PMC article. - Aging increases stromal/osteoblastic cell-induced osteoclastogenesis and alters the osteoclast precursor pool in the mouse.
Cao JJ, Wronski TJ, Iwaniec U, Phleger L, Kurimoto P, Boudignon B, Halloran BP. Cao JJ, et al. J Bone Miner Res. 2005 Sep;20(9):1659-68. doi: 10.1359/JBMR.050503. Epub 2005 May 2. J Bone Miner Res. 2005. PMID: 16059637 - The dendritic cell-specific transmembrane protein DC-STAMP is essential for osteoclast fusion and osteoclast bone-resorbing activity.
Miyamoto T. Miyamoto T. Mod Rheumatol. 2006;16(6):341-2. doi: 10.1007/s10165-006-0524-0. Epub 2006 Dec 20. Mod Rheumatol. 2006. PMID: 17164993 Review. - Regulators of osteoclast differentiation and cell-cell fusion.
Miyamoto T. Miyamoto T. Keio J Med. 2011;60(4):101-5. doi: 10.2302/kjm.60.101. Keio J Med. 2011. PMID: 22200633 Review.
Cited by
- Antiresorptive activity of bacillus-fermented antler extracts: inhibition of osteoclast differentiation.
Choi SW, Moon SH, Yang HJ, Kwon DY, Son YJ, Yu R, Kim YS, Kim SI, Chae EJ, Park SJ, Kim SH. Choi SW, et al. Evid Based Complement Alternat Med. 2013;2013:748687. doi: 10.1155/2013/748687. Epub 2013 Feb 10. Evid Based Complement Alternat Med. 2013. PMID: 23509596 Free PMC article. - Osteoclast Multinucleation: Review of Current Literature.
Kodama J, Kaito T. Kodama J, et al. Int J Mol Sci. 2020 Aug 8;21(16):5685. doi: 10.3390/ijms21165685. Int J Mol Sci. 2020. PMID: 32784443 Free PMC article. Review. - Deficiency of Lipin2 Results in Enhanced NF-κB Signaling and Osteoclast Formation in RAW-D Murine Macrophages.
Watahiki A, Hoshikawa S, Chiba M, Egusa H, Fukumoto S, Inuzuka H. Watahiki A, et al. Int J Mol Sci. 2021 Mar 12;22(6):2893. doi: 10.3390/ijms22062893. Int J Mol Sci. 2021. PMID: 33809261 Free PMC article. - Osteoclast fusion and regulation by RANKL-dependent and independent factors.
Xing L, Xiu Y, Boyce BF. Xing L, et al. World J Orthop. 2012 Dec 18;3(12):212-22. doi: 10.5312/wjo.v3.i12.212. World J Orthop. 2012. PMID: 23362465 Free PMC article. - The PPAR-γ antagonist T007 inhibits RANKL-induced osteoclastogenesis and counteracts OVX-induced bone loss in mice.
Li X, Ning L, Ma J, Xie Z, Zhao X, Wang G, Wan X, Qiu P, Yao T, Wang H, Fan S, Wan S. Li X, et al. Cell Commun Signal. 2019 Oct 26;17(1):136. doi: 10.1186/s12964-019-0442-3. Cell Commun Signal. 2019. PMID: 31655621 Free PMC article.
References
- Roodman, G.D. 1999. Cell biology of the osteoclast. Exp. Hematol. 27:1229–1241. - PubMed
- Suda, T., N. Takahashi, N. Udagawa, E. Jimi, M.T. Gillespie, and T.J. Martin. 1999. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 20:345–357. - PubMed
- Boyle, W.J., W.S. Simonet, and D.L. Lacey. 2003. Osteoclast differentiation and activation. Nature. 423:337–342. - PubMed
- Teitelbaum, S.L., and F.P. Ross. 2003. Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 4:638–649. - PubMed
- Han, X., H. Sterling, Y. Chen, C. Saginario, E.J. Brown, W.A. Frazier, F.P. Lindberg, and A. Vignery. 2000. CD47, a ligand for the macrophage fusion receptor, participates in macrophage multinucleation. J. Biol. Chem. 275:37984–37992. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials