Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2 - PubMed (original) (raw)
Comparative Study
Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2
Wenhui Li et al. J Virol. 2004 Oct.
Abstract
Replication of viruses in species other than their natural hosts is frequently limited by entry and postentry barriers. The coronavirus that causes severe acute respiratory syndrome (SARS-CoV) utilizes the receptor angiotensin-converting enzyme 2 (ACE2) to infect cells. Here we compare human, mouse, and rat ACE2 molecules for their ability to serve as receptors for SARS-CoV. We found that, compared to human ACE2, murine ACE2 less efficiently bound the S1 domain of SARS-CoV and supported less-efficient S protein-mediated infection. Rat ACE2 was even less efficient, at near background levels for both activities. Murine 3T3 cells expressing human ACE2 supported SARS-CoV replication, whereas replication was less than 10% as efficient in the same cells expressing murine ACE2. These data imply that a mouse transgenically expressing human ACE2 may be a useful animal model of SARS.
Figures
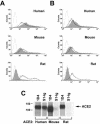
FIG. 1.
SARS-CoV S1 domain binds rodent ACE2 less efficiently than human ACE2. (A) 293T cells transfected with 10 μg of plasmid encoding N-terminally Myc-tagged ACE2 of the indicated species were stained with the anti-Myc-tag antibody 9E10 (heavy line), with S1-Ig (a fusion of the SARS-CoV S1 domain with the Fc domain of human IgG1 [shaded area]), or with secondary antibody alone (light line), and analyzed by flow cytometry. (B) Same as for panel A, except that 5 μg of plasmid was used for transfection. (C) 293T cells transfected with plasmids encoding C-terminally tagged ACE2 of the indicated species were metabolically labeled with [35S]cysteine and [35S]methionine and lysed as previously described (20). Cell lysates were immunoprecipitated with either the antitag antibody 1D4 or S1-Ig and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
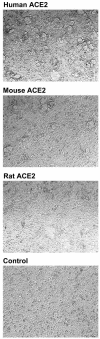
FIG. 2.
Rodent ACE2 mediates less-efficient syncytial formation with S protein-expressing cells than human ACE2. 293T cells transfected with plasmids encoding ACE2 of the indicated species or with vector alone (control) were mixed at a 1:1 ratio with 293T cells transfected with a plasmid encoding a codon-optimized form of a SARS-CoV S protein. Multinucleated syncytia were observed 48 h after the cells were mixed. The experiment shown is representative of three experiments with similar results.
FIG. 3.
Infection with S protein-pseudotyped retrovirus is less efficient in cells expressing rodent ACE2 than in those expressing human ACE2. (A) An aliquot of cells analyzed as shown in Fig. 1B was incubated with S protein-pseudotyped SIV-GFP. After 48 h, the cells were harvested and analyzed by flow cytometry for GFP expression. GFP levels indicating infection (dark gray) are compared with anti-Myc antibody (light gray) and S1-Ig (medium gray) recognition in the same cells. (B) Same experiment as shown in panel A except that cells analyzed as shown in Fig. 1A were infected with SIV-GFP. (C) Typical image of GFP expression of cells transfected and infected as shown in panel B. The figure is representative of two similar experiments with similar results.
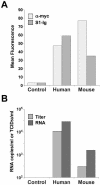
FIG. 4.
Replication of SARS-CoV in murine cells is enhanced by human receptor. (A) Murine 3T3 cells were transfected with plasmid encoding Myc-tagged variants of human or mouse ACE2 as indicated, or with vector alone (control), and analyzed by flow cytometry using the anti-Myc-tag antibody 9E10 (light gray bars) or S1-Ig (medium gray bars). (B) An aliquot of the cells analyzed as shown in panel A was incubated with SARS-CoV for 4 days. Peak viral titers (medium gray) were determined by measuring the cytopathicity in Vero E6 cells incubated with cell supernatants as previously described (13). Viral RNA levels (dark gray) were determined by semiquantitative reverse transcription-PCR of cell supernatants, as previously described (13). Viral titers and RNA levels of control cells were below 100 50% tissue culture infective doses (TCID50) per ml and 100 copies per ml, the limits of detection in both assays. The figure shows an experiment representative of two experiments with similar results.
Similar articles
- Rhesus angiotensin converting enzyme 2 supports entry of severe acute respiratory syndrome coronavirus in Chinese macaques.
Chen Y, Liu L, Wei Q, Zhu H, Jiang H, Tu X, Qin C, Chen Z. Chen Y, et al. Virology. 2008 Nov 10;381(1):89-97. doi: 10.1016/j.virol.2008.08.016. Epub 2008 Sep 17. Virology. 2008. PMID: 18801550 Free PMC article. - Angiotensin-converting enzyme 2 (ACE2) from raccoon dog can serve as an efficient receptor for the spike protein of severe acute respiratory syndrome coronavirus.
Xu L, Zhang Y, Liu Y, Chen Z, Deng H, Ma Z, Wang H, Hu Z, Deng F. Xu L, et al. J Gen Virol. 2009 Nov;90(Pt 11):2695-2703. doi: 10.1099/vir.0.013490-0. Epub 2009 Jul 22. J Gen Virol. 2009. PMID: 19625462 - TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein.
Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pöhlmann S. Heurich A, et al. J Virol. 2014 Jan;88(2):1293-307. doi: 10.1128/JVI.02202-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227843 Free PMC article. - SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium.
Sims AC, Burkett SE, Yount B, Pickles RJ. Sims AC, et al. Virus Res. 2008 Apr;133(1):33-44. doi: 10.1016/j.virusres.2007.03.013. Epub 2007 Apr 23. Virus Res. 2008. PMID: 17451829 Free PMC article. Review. - Severe acute respiratory syndrome coronavirus entry as a target of antiviral therapies.
Kuhn JH, Li W, Radoshitzky SR, Choe H, Farzan M. Kuhn JH, et al. Antivir Ther. 2007;12(4 Pt B):639-50. Antivir Ther. 2007. PMID: 17944271 Review.
Cited by
- Broad and Differential Animal Angiotensin-Converting Enzyme 2 Receptor Usage by SARS-CoV-2.
Zhao X, Chen D, Szabla R, Zheng M, Li G, Du P, Zheng S, Li X, Song C, Li R, Guo JT, Junop M, Zeng H, Lin H. Zhao X, et al. J Virol. 2020 Aug 31;94(18):e00940-20. doi: 10.1128/JVI.00940-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32661139 Free PMC article. - Host Receptors of Influenza Viruses and Coronaviruses-Molecular Mechanisms of Recognition.
Sriwilaijaroen N, Suzuki Y. Sriwilaijaroen N, et al. Vaccines (Basel). 2020 Oct 6;8(4):587. doi: 10.3390/vaccines8040587. Vaccines (Basel). 2020. PMID: 33036202 Free PMC article. Review. - Interactions between SARS coronavirus and its receptor.
Li F, Li W, Farzan M, Harrison SC. Li F, et al. Adv Exp Med Biol. 2006;581:229-34. doi: 10.1007/978-0-387-33012-9_38. Adv Exp Med Biol. 2006. PMID: 17037534 Free PMC article. No abstract available. - Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus.
McCray PB Jr, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S. McCray PB Jr, et al. J Virol. 2007 Jan;81(2):813-21. doi: 10.1128/JVI.02012-06. Epub 2006 Nov 1. J Virol. 2007. PMID: 17079315 Free PMC article. - Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs.
Sims AC, Baric RS, Yount B, Burkett SE, Collins PL, Pickles RJ. Sims AC, et al. J Virol. 2005 Dec;79(24):15511-24. doi: 10.1128/JVI.79.24.15511-15524.2005. J Virol. 2005. PMID: 16306622 Free PMC article.
References
- Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. - PubMed
- Farzan, M., T. Mirzabekov, P. Kolchinsky, R. Wyatt, M. Cayabyab, N. P. Gerard, C. Gerard, J. Sodroski, and H. Choe. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667-676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous