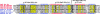Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity - PubMed (original) (raw)
Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity
Tim A Rand et al. Proc Natl Acad Sci U S A. 2004.
Abstract
RNA interference is carried out by the small double-stranded RNA-induced silencing complex (RISC). The RISC-bound small RNA guides the RISC complex to identify and cleave mRNAs with complementary sequences. The proteins that make up the RISC complex and cleave mRNA have not been unequivocally defined. Here, we report the biochemical purification of RISC activity to homogeneity from Drosophila Schnieder 2 cell extracts. Argonaute 2 (Ago-2) is the sole protein component present in the purified, functional RISC. By using a bioinformatics method that combines sequence-profile analysis with predicted protein secondary structure, we found homology between the PIWI domain of Ago-2 and endonuclease V and identified potential active-site amino acid residues within the PIWI domain of Ago-2.
Figures
Fig. 1.
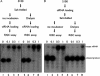
RISC assembly is sensitive, but preassembled RISC is resistant to high salt. S2 S100 was loaded in vitro with siRNA to program RISC to cleave a G-cap radiolabeled mRNA target (see Materials and Methods). (A) We added 0, 0.1, 0.3, or 1 M NaCl to extract, and the samples were either incubated on ice (lanes 1-4) or dialyzed back to starting buffer conditions at 4°C for 4 h (lanes 5-8) before siRNA and 1 mM ATP was introduced. After a 30-min incubation at room temperature, radiolabeled target mRNA was added, and the reactions were incubated for an additional 30 min at 30°C to measure RISC activity. (B) The indicated concentration of salt was added after siRNA loading. The samples were placed on ice (lanes 1-4) or dialyzed (in buffer A) (lanes 5-8) for 4 h at 4°C. All samples were then tested for RISC (as described above).
Fig. 2.
Purification of preassembled RISC activity. (A) Schematic representation of the purification procedure. (B) RISC assay with individual fractions from the phenyl-Sepharose column (17-29). The first lane shows starting material activity. All samples were dialyzed to 200 mM KOAc before being assayed for activity (because high salt masks activity). (C) The cleaved mRNA product after incubation with the streptavidin-coated magnetic beads after detergent and salt washes (see Materials and Methods for details). Upper is from a parallel purification in which the loaded siRNA was not biotinylated. Lower shows the biotinylated siRNA pull-down activity. Lanes 1 and 3 show the activity before and after the high-salt wash, respectively. Lane 2 shows the activity that is left in the supernatant after the pull down. (D) The corresponding proteins identified from MS of the trypsinized beads. Under the most stringent conditions (biotin tag, detergent wash, and salt wash), only Ago-2 was detected (see Fig. 3 for peptide list).
Fig. 3.
Peptides recovered from the RISC purification. The 33 peptides recovered all map to Ago-2 (GenBank accession no. 24664664; full-length Ago-2) and cover a significant portion of the entire protein. Three nonredundant peptides had amino acids that were different from GenBank entry 24664664 but that agree with the sequence in GenBank entry 20151489 (likely an Ago-2 allelic variant fragment). The amino acid detected in our data (taken from S2 cells) is placed above the 24664664 sequence for each case. Bars indicate the best match sequence for each collected MS signal.
Fig. 4.
Alignment of PIWI domains with selected endonucleases. Ago-2 PIWI domains from D. melanogaster (Dm) and Mus musculus (Mm) are aligned with PIWI domain from P. furiosus (Pf) and endonucleases V, UVRC and RNaseHI from E. coli (Ec). GenBank accession nos. are given at the end of sequence names. Secondary-structure prediction for Ago-2_Dm and secondary structure of RNaseHI (PDB ID 2RN2) are given above and below the alignment, respectively. The β-strands and α-helices are indicated by E and H, respectively. Functionally important conserved residues are shown in white letters boxed in black, uncharged residues at mainly hydrophobic positions are highlighted in yellow, and conserved small residues are highlighted in gray. The first and last residue numbers of the shown sequences are indicated before and after each sequence. Some nonconserved residues are omitted, and the number of omitted residues is shown in parentheses.
Similar articles
- [Components and assembly of RNA-induced silencing complex].
Song XM, Yan F, Du LX. Song XM, et al. Yi Chuan. 2006 Jun;28(6):761-6. Yi Chuan. 2006. PMID: 16818443 Review. Chinese. - Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes.
Kim K, Lee YS, Carthew RW. Kim K, et al. RNA. 2007 Jan;13(1):22-9. doi: 10.1261/rna.283207. Epub 2006 Nov 22. RNA. 2007. PMID: 17123955 Free PMC article. - Rapid and specific purification of Argonaute-small RNA complexes from crude cell lysates.
Flores-Jasso CF, Salomon WE, Zamore PD. Flores-Jasso CF, et al. RNA. 2013 Feb;19(2):271-9. doi: 10.1261/rna.036921.112. Epub 2012 Dec 18. RNA. 2013. PMID: 23249751 Free PMC article. - Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway.
Pham JW, Sontheimer EJ. Pham JW, et al. J Biol Chem. 2005 Nov 25;280(47):39278-83. doi: 10.1074/jbc.M509202200. Epub 2005 Sep 22. J Biol Chem. 2005. PMID: 16179342 - RISC assembly: Coordination between small RNAs and Argonaute proteins.
Kobayashi H, Tomari Y. Kobayashi H, et al. Biochim Biophys Acta. 2016 Jan;1859(1):71-81. doi: 10.1016/j.bbagrm.2015.08.007. Epub 2015 Aug 22. Biochim Biophys Acta. 2016. PMID: 26303205 Review.
Cited by
- Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway.
Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Ghildiyal M, et al. RNA. 2010 Jan;16(1):43-56. doi: 10.1261/rna.1972910. Epub 2009 Nov 16. RNA. 2010. PMID: 19917635 Free PMC article. - Statistical use of argonaute expression and RISC assembly in microRNA target identification.
Stanhope SA, Sengupta S, den Boon J, Ahlquist P, Newton MA. Stanhope SA, et al. PLoS Comput Biol. 2009 Sep;5(9):e1000516. doi: 10.1371/journal.pcbi.1000516. Epub 2009 Sep 25. PLoS Comput Biol. 2009. PMID: 19779550 Free PMC article. - Mosquito antiviral defense mechanisms: a delicate balance between innate immunity and persistent viral infection.
Lee WS, Webster JA, Madzokere ET, Stephenson EB, Herrero LJ. Lee WS, et al. Parasit Vectors. 2019 Apr 11;12(1):165. doi: 10.1186/s13071-019-3433-8. Parasit Vectors. 2019. PMID: 30975197 Free PMC article. Review. - Nucleus-specific RNAi nanoplatform for targeted regulation of nuclear lncRNA function and effective cancer therapy.
Huang Z, Liu S, Lu N, Xu L, Shen Q, Huang Z, Huang Z, Saw PE, Xu X. Huang Z, et al. Exploration (Beijing). 2022 Jul 26;2(5):20220013. doi: 10.1002/EXP.20220013. eCollection 2022 Oct. Exploration (Beijing). 2022. PMID: 37325502 Free PMC article. - Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1.
Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Förstemann K, et al. Cell. 2007 Jul 27;130(2):287-97. doi: 10.1016/j.cell.2007.05.056. Cell. 2007. PMID: 17662943 Free PMC article.
References
- Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806-811. - PubMed
- Liu, Q., Rand, T. A., Kalidas, S., Du, F., Kim, H. E., Smith, D. P. & Wang, X. (2003) Science 301, 1921-1925. - PubMed
- Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363-366. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases