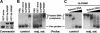The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin - PubMed (original) (raw)
The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin
Taiping Chen et al. Mol Cell Biol. 2004 Oct.
Abstract
Dnmt3a and Dnmt3b are responsible for the establishment of DNA methylation patterns during development. These proteins contain, in addition to a C-terminal catalytic domain, a unique N-terminal regulatory region that harbors conserved domains, including a PWWP domain. The PWWP domain, characterized by the presence of a highly conserved proline-tryptophan-tryptophan-proline motif, is a module of 100 to 150 amino acids found in many chromatin-associated proteins. However, the function of the PWWP domain remains largely unknown. In this study, we provide evidence that the PWWP domains of Dnmt3a and Dnmt3b are involved in functional specialization of these enzymes. We show that both endogenous and green fluorescent protein-tagged Dnmt3a and Dnmt3b are particularly concentrated in pericentric heterochromatin. Mutagenesis analysis indicates that their PWWP domains are required for their association with pericentric heterochromatin. Disruption of the PWWP domain abolishes the ability of Dnmt3a and Dnmt3b to methylate the major satellite repeats at pericentric heterochromatin. Furthermore, we demonstrate that the Dnmt3a PWWP domain has little DNA-binding ability, in contrast to the Dnmt3b PWWP domain, which binds DNA nonspecifically. Collectively, our results suggest that the PWWP domains of Dnmt3a and Dnmt3b are essential for targeting these enzymes to pericentric heterochromatin, probably via a mechanism other than protein-DNA interactions.
Figures
FIG. 1.
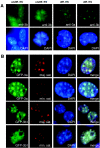
Dnmt3a and Dnmt3b are enriched in pericentric heterochromatin. (A) Localization of endogenous Dnmt3a and Dnmt3b. Undifferentiated and differentiated ES cells were fixed, permeabilized, and immunostained with rabbit polyclonal anti-Dnmt3a or anti-Dnmt3b, followed by fluorescein isothiocyanate-conjugated goat anti-rabbit secondary antibody. The top panel shows the fluorescein isothiocyanate signals, and the bottom panel shows the nuclei stained with DAPI. (B) Colocalization of Dnmt3a and Dnmt3b with the major satellite repeats. GFP-Dnmt3a and GFP-Dnmt3b1 were individually transfected into NIH 3T3 cells, and the cells were fixed and analyzed by FISH with indocarbocyanine-labeled oligonucleotide probes specific for mouse major or minor satellite repeats. The signals for GFP, indocarbocyanine (Cy3), and DAPI as well as the merged images are shown.
FIG. 2.
PWWP domain is required for preferential localization of Dnmt3a and Dnmt3b to pericentric heterochromatin. (A) The GFP-Dnmt3a and GFP-Dnmt3b constructs were transfected into NIH 3T3 cells, and the protein localization patterns were analyzed by fluorescence microscopy. The typical localization patterns observed are shown: punctate nuclear localization with accumulation in pericentric heterochromatin (pattern A), punctate nuclear localization without accumulation in pericentric heterochromatin (pattern B), diffuse nuclear localization with accumulation in pericentric heterochromatin (pattern C), diffuse nuclear localization without accumulation in pericentric heterochromatin (pattern D), and diffuse nuclear and cytoplasmic localization (pattern E). (B and C) Schematic diagrams of the GFP-Dnmt3a and GFP-Dnmt3b constructs and quantitation of different localization patterns. In the diagrams, the conserved PWWP and ATRX homology domains, the methyltransferase motifs (I, IV, VI, IX, and X), the putative NLSs, and the sites of alternative splicing are indicated. The positions of deletions and point mutations are indicated by horizontal and vertical thin lines, respectively. At the left, Δ indicates a deletion, → denotes amino acid substitutions, and the numbers are amino acid numbers. The GFP moiety (not shown) was fused to the N termini of the Dnmt3a and Dnmt3b proteins. For each construct, an average of 200 to 300 transfected (green) cells from two separate experiments were counted, and their localization patterns are shown as percentages.
FIG. 3.
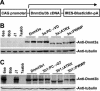
Stable expression of Dnmt3a and Dnmt3b proteins in _Dnmt3a_−/− _Dnmt3b_−/− ES cells. cDNAs encoding wild-type or mutant Dnmt3a and Dnmt3b proteins were subcloned in a bicistronic expression vector (schematically shown in A), and these constructs were individually electroporated into late-passage (P50) _Dnmt3a_−/− _Dnmt3b_−/− double mutant (7aabb) ES cells, which were subsequently selected in blasticidin-containing medium for 7 days. Blasticidin-resistant clones as well as wild-type (J1), _Dnmt3a_−/− (6aa), _Dnmt3b_−/− (8bb), and 7aabb ES cells were analyzed with immunoblotting with anti-Dnmt3a (B) or anti-Dnmt3b (C). As a loading control, the same membranes were immunoblotted with antitubulin.
FIG. 4.
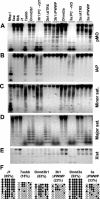
PWWP domain is specifically required for Dnmt3a and Dnmt3b to restore methylation of the major satellite repeats. (A-E) Methylation analysis by Southern hybridization. Genomic DNA from the indicated ES cell lines was digested with HpaII (A-C), Mae II (D), or EcoRV plus HhaI (E) and hybridized to probes for endogenous C-type retrovirus repeats (pMO) (A), the intracisternal A particle repeats (B), the minor satellite repeats (C), the major satellite repeats (D), or the 5′ region of Xist (E). DNA from J1 cells digested with MspI was used as a control for complete digestion. (F) Analysis of the methylation status of the major satellite repeating unit by bisulfite sequencing. Genomic DNA from J1 and 7aabb cells as well as stable cell lines expressing Dnmt3a, Dnmt3aΔPWWP, Dnmt3b1, and Dnmt3b1ΔPWWP was analyzed. The methylation status of six CpG sites from 9 to 12 individual clones is shown schematically (black circles represent methylated sites), and the percentages of methylated CpG sites are indicated in parentheses.
FIG. 5.
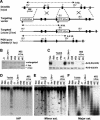
Targeted deletion of Dnmt3b exon 7 results in demethylation of the major satellite repeats. (A) Strategy for targeted deletion of Dnmt3b exon 7. The top line shows the Dnmt3b genomic structure with exons represented by vertical bars. The targeting vector (second line) was constructed by replacing exon 7 with a PGK-puromycin cassette flanked by loxP sites (shown as triangles). A PGK-DTA cassette was introduced for negative selection to increase the targeting frequency. The third line represents the mutant locus resulting from homologous recombination between the targeting vector and the wild-type locus (2 lox). The fourth line represents the targeted locus with the PGK-puromycin cassette deleted by Cre (1 lox). (B) Southern analysis of the genotype of ES cell lines. Genomic DNA was digested with EcoRV and hybridized to a 5′ external probe, as shown in A. The 6.0-kb untargeted allele and the 4.0-kb (2 lox) and 4.4-kb (1 lox) targeted alleles are indicated. (C) Lysates from the indicated cell lines were immunoblotted with anti-Dnmt3b (top) and antitubulin (bottom) antibodies. (D-F) Genomic DNA from the indicated ES cell lines was digested with HpaII (D and E) or Mae II (F) and hybridized to probes for the intracisternal A particle repeats (D), the minor satellite repeats (E), or the major satellite repeats (F).
FIG. 6.
PWWP domains of Dnmt3a and Dnmt3b have different DNA-binding abilities. (A) GST fusion proteins stained with Coomassie blue. (B) Electrophoretic mobility shift assay. 32P-labeled DNA probes corresponding to the 234-bp unit sequence of the major satellite repeats or similar-sized random genomic sequences (control) were incubated with phosphate-buffered saline (PBS) or GST fusion proteins (1 μg), and the reactions were analyzed by native acrylamide gel electrophoresis and autoradiography. (C) GST-Dnmt3b:PWWP (1 μg) was used in the presence of increasing amounts (0, 2, 10, 30, and 100 ng) of sheared salmon sperm DNA.
Similar articles
- DNMT3B PWWP mutations cause hypermethylation of heterochromatin.
Taglini F, Kafetzopoulos I, Rolls W, Musialik KI, Lee HY, Zhang Y, Marenda M, Kerr L, Finan H, Rubio-Ramon C, Gautier P, Wapenaar H, Kumar D, Davidson-Smith H, Wills J, Murphy LC, Wheeler A, Wilson MD, Sproul D. Taglini F, et al. EMBO Rep. 2024 Mar;25(3):1130-1155. doi: 10.1038/s44319-024-00061-5. Epub 2024 Jan 30. EMBO Rep. 2024. PMID: 38291337 Free PMC article. - Chromatin targeting of de novo DNA methyltransferases by the PWWP domain.
Ge YZ, Pu MT, Gowher H, Wu HP, Ding JP, Jeltsch A, Xu GL. Ge YZ, et al. J Biol Chem. 2004 Jun 11;279(24):25447-54. doi: 10.1074/jbc.M312296200. Epub 2004 Mar 3. J Biol Chem. 2004. PMID: 14998998 - The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds.
Qiu C, Sawada K, Zhang X, Cheng X. Qiu C, et al. Nat Struct Biol. 2002 Mar;9(3):217-24. doi: 10.1038/nsb759. Nat Struct Biol. 2002. PMID: 11836534 Free PMC article. - Using human disease mutations to understand de novo DNA methyltransferase function.
Rolls W, Wilson MD, Sproul D. Rolls W, et al. Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review. - Tissue-specific roles of de novo DNA methyltransferases.
Tóth DM, Szeri F, Ashaber M, Muazu M, Székvölgyi L, Arányi T. Tóth DM, et al. Epigenetics Chromatin. 2025 Jan 17;18(1):5. doi: 10.1186/s13072-024-00566-2. Epigenetics Chromatin. 2025. PMID: 39819598 Free PMC article. Review.
Cited by
- Potential advantages of DNA methyltransferase 1 (DNMT1)-targeted inhibition for cancer therapy.
Jung Y, Park J, Kim TY, Park JH, Jong HS, Im SA, Robertson KD, Bang YJ, Kim TY. Jung Y, et al. J Mol Med (Berl). 2007 Oct;85(10):1137-48. doi: 10.1007/s00109-007-0216-z. Epub 2007 Jun 15. J Mol Med (Berl). 2007. PMID: 17571247 - Screening for genes that accelerate the epigenetic aging clock in humans reveals a role for the H3K36 methyltransferase NSD1.
Martin-Herranz DE, Aref-Eshghi E, Bonder MJ, Stubbs TM, Choufani S, Weksberg R, Stegle O, Sadikovic B, Reik W, Thornton JM. Martin-Herranz DE, et al. Genome Biol. 2019 Aug 14;20(1):146. doi: 10.1186/s13059-019-1753-9. Genome Biol. 2019. PMID: 31409373 Free PMC article. - Suppression of intestinal neoplasia by deletion of Dnmt3b.
Lin H, Yamada Y, Nguyen S, Linhart H, Jackson-Grusby L, Meissner A, Meletis K, Lo G, Jaenisch R. Lin H, et al. Mol Cell Biol. 2006 Apr;26(8):2976-83. doi: 10.1128/MCB.26.8.2976-2983.2006. Mol Cell Biol. 2006. PMID: 16581773 Free PMC article. - PWWP domains and their modes of sensing DNA and histone methylated lysines.
Rona GB, Eleutherio ECA, Pinheiro AS. Rona GB, et al. Biophys Rev. 2016 Mar;8(1):63-74. doi: 10.1007/s12551-015-0190-6. Epub 2016 Jan 14. Biophys Rev. 2016. PMID: 28510146 Free PMC article. Review. - DNA Methylation in T-Cell Development and Differentiation.
Correa LO, Jordan MS, Carty SA. Correa LO, et al. Crit Rev Immunol. 2020;40(2):135-156. doi: 10.1615/CritRevImmunol.2020033728. Crit Rev Immunol. 2020. PMID: 32749092 Free PMC article. Review.
References
- Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. - PubMed
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. - PubMed
- Chen, T., and E. Li. 2004. Structure and function of eukaryotic DNA methyltransferases. Curr. Top. Dev. Biol. 60:55-89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases