Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3 - PubMed (original) (raw)
. 2004 Oct 27;23(21):4286-96.
doi: 10.1038/sj.emboj.7600430. Epub 2004 Sep 30.
David Shultis, Zuzana Jasencakova, Jörg Fuchs, Lianna Johnson, Daniel Schubert, Debasis Patnaik, Sriharsa Pradhan, Justin Goodrich, Ingo Schubert, Thomas Jenuwein, Sepideh Khorasanizadeh, Steven E Jacobsen
Affiliations
- PMID: 15457214
- PMCID: PMC524394
- DOI: 10.1038/sj.emboj.7600430
Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3
Anders M Lindroth et al. EMBO J. 2004.
Erratum in
- EMBO J. 2011 May 4;30(9):1874
Abstract
Both DNA methylation and post-translational histone modifications contribute to gene silencing, but the mechanistic relationship between these epigenetic marks is unclear. Mutations in two Arabidopsis genes, the KRYPTONITE (KYP) histone H3 lysine 9 (H3K9) methyltransferase and the CHROMOMETHYLASE3 (CMT3) DNA methyltransferase, cause a reduction of CNG DNA methylation, suggesting that H3K9 methylation controls CNG DNA methylation. Here we show that the chromodomain of CMT3 can directly interact with the N-terminal tail of histone H3, but only when it is simultaneously methylated at both the H3K9 and H3K27 positions. Furthermore, using chromatin immunoprecipitation analysis and immunohistolocalization experiments, we found that H3K27 methylation colocalizes with H3K9 methylation at CMT3-controlled loci. The H3K27 methylation present at heterochromatin was not affected by mutations in KYP or in several Arabidopsis PcG related genes including the Enhancer of Zeste homologs, suggesting that a novel pathway controls heterochromatic H3K27 methylation. Our results suggest a model in which H3K9 methylation by KYP, and H3K27 methylation by an unknown enzyme provide a combinatorial histone code for the recruitment of CMT3 to silent loci.
Figures
Figure 1
lhp1 mutants do not affect DNA methylation or gene silencing in heterochromatin. (A–C) Southern blot analysis with the DNA methylation-sensitive endonuclease _Cfo_I and _Hpa_II for CG sites, and _Bgl_II and _Msp_I for CNG sites, probed with centromeric 180 bp repeat (CEN), Ta3, and FWA. _fwa_-1 is a mutant lacking methylation of the FWA gene. cmt3-7 (included as a control) reduces CNG methylation, and therefore shows a downshifted banding pattern. (D) Semiquantitative RT–PCR showing reactivation of retroelement sequences in cmt3 but not in lhp1 mutants. (E) Immunohistochemical staining of wild-type and lhp1 interphase nuclei with dimethyl H3K9 antibodies preferentially labeling heterochromatic chromocenters. The lhp1-4 mutant does not affect distribution of methyl H3K9. (F) Floral phenotype of the clark kent-st (clk-st) lhp1-4 double mutant. The left flower, a clk-st mutant, shows supernumerary stamens and a defective gynoecium. The middle flower, an lhp1-4 mutant, shows a terminal flower with three normal gynoecia. The right flower, a clk-st lhp1-4 double mutant, shows a compound terminal flower containing two _clk-st_-like defective gynoecia.
Figure 2
Interaction of the CMT3 chromodomain with the N-terminal tail of histone H3. (A) Domain composition of CMT3. Roman numerals denote the conserved motifs within the catalytic DNA methyltransferase region. (B) Alignment of the chromodomains of CMT3 and ZMET2 (a functional CMT gene from Zea mays; Papa et al, 2001) proteins with those of the HP1, LHP1, and Pc proteins. Secondary structure elements drawn above the sequence correspond to the structure of the HP1 chromodomain. Positions F382, W409, and Y412, highlighted in red, show the sites of conserved aromatic residues that form an aromatic cage. (C) Fluorescence polarization-based binding results for the interaction of the CMT3 chromodomain with H3 peptides. The CMT3 chromodomain binds to a doubly methylated H3K9/K27 peptide, but not to either the unmodified or singly methylated peptides. Binding was eliminated by mutation of the conserved aromatic residue F382 to alanine. (D) Binding of CMT3 chromodomain to methyl H3K9/K27 as measured by ITC. The solid line corresponds to the fit of the integrated heats of injections to an interaction stoichiometry of 0.5; see the inset for the thermodynamic parameters of the best fit of the data using the MicroCal software.
Figure 3
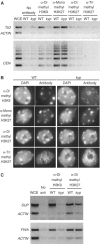
Colocalization of methyl H3K9 and H3K27 at heterochromatin. (A) ChIP analysis showing methyl H3K9 and H3K27 enrichment at the Ta3 retrotransposon and CEN repeats, relative to ACTIN. kyp affects levels of methyl H3K9 but not mono-, di-, or trimethyl H3K27. ACTIN is used for normalization, and the amount of chromatin used in each sample (for both the Ta3 and CEN experiments) was adjusted so that an equal amount of ACTIN was amplified. WCE, whole-cell extract. (B) Distribution of mono-, di-, and trimethyl H3K27 in interphase leaf nuclei. Mono- and dimethyl H3K27 are enriched at DAPI-stained chromocenters, while trimethyl H3K27 signals are more evenly distributed. kyp reduces dimethyl H3K9 at heterochromatin, but not H3K27 methylation. (C) ChIP analysis showing enrichment of both H3K9 and H3K27 methylation at SUPERMAN and FWA relative to ACTIN.
Figure 4
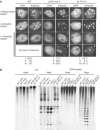
Effect of PcG mutations on the distribution of methyl H3K27 in leaf interphase nuclei and on DNA methylation. (A) Distribution of mono-, di-, and trimethyl H3K27 in the clf-50 swn-3 double mutant and the fie TK114 mutant. These mutants do not reduce heterochromatic H3K27 methylation, but do reduce euchromatic H3K27m2. Whereas H3K27m3 was evenly distributed in wild-type nuclei, in a fraction of clf-50 swn-3 and fie TK114 nuclei, signal was concentrated in chromocenters. The percent of nuclei showing three different staining patterns with the H3K27m3 antibody is listed below (types I, II, and III). Type I is a pattern similar to wild type with signals evenly distributed within nuclei. Type II is H3K27m3 signals concentrated at chromocenters (examples shown in the lower row of photographs). Type III is an intermediate pattern where H3K27m3 was concentrated at only those chromocenters associating with nucleoli (not shown). At least 150 nuclei were evaluated for each genotype. (B) Southern blot analysis showing the lack of effect of the clf-50 swn-3 double mutant and the fie TK114 mutant on DNA methylation. The cmt3 and kyp mutants are included as controls and show loss of CNG methylation (an increased band intensity of the low-molecular-weight bands as compared to the high-molecular-weight bands). Two wild-type controls are shown because the cmt3 and kyp mutants are in the L_er_ background, and the clf-50 swn-3 and fie TK114 mutants are in the Col background.
Figure 5
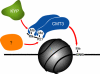
Model for the relationship of histone methylation and CNG DNA methylation. Histone H3K9 methylation through KRYPTONITE and H3K27 methylation through an unknown methyltransferase act to target the activity of the CMT3 DNA methyltransferase.
Similar articles
- The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase.
Fuks F, Hurd PJ, Deplus R, Kouzarides T. Fuks F, et al. Nucleic Acids Res. 2003 May 1;31(9):2305-12. doi: 10.1093/nar/gkg332. Nucleic Acids Res. 2003. PMID: 12711675 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Single-genome analysis reveals a heterogeneous association of the herpes simplex virus genome with H3K27me2 and the reader PHF20L1 following infection of human fibroblasts.
Francois AK, Rohani A, Loftus M, Dochnal S, Hrit J, McFarlane S, Whitford A, Lewis A, Krakowiak P, Boutell C, Rothbart SB, Kashatus D, Cliffe AR. Francois AK, et al. mBio. 2024 Apr 10;15(4):e0327823. doi: 10.1128/mbio.03278-23. Epub 2024 Feb 27. mBio. 2024. PMID: 38411116 Free PMC article. - Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.
Elwenspoek MM, Thom H, Sheppard AL, Keeney E, O'Donnell R, Jackson J, Roadevin C, Dawson S, Lane D, Stubbs J, Everitt H, Watson JC, Hay AD, Gillett P, Robins G, Jones HE, Mallett S, Whiting PF. Elwenspoek MM, et al. Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article. - The effectiveness of abstinence-based and harm reduction-based interventions in reducing problematic substance use in adults who are experiencing homelessness in high income countries: A systematic review and meta-analysis: A systematic review.
O'Leary C, Ralphs R, Stevenson J, Smith A, Harrison J, Kiss Z, Armitage H. O'Leary C, et al. Campbell Syst Rev. 2024 Apr 21;20(2):e1396. doi: 10.1002/cl2.1396. eCollection 2024 Jun. Campbell Syst Rev. 2024. PMID: 38645303 Free PMC article. Review.
Cited by
- Evolutional heterochromatin condensation delineates chromocenter formation and retrotransposon silencing in plants.
Zhang W, Cheng L, Li K, Xie L, Ji J, Lei X, Jiang A, Chen C, Li H, Li P, Sun Q. Zhang W, et al. Nat Plants. 2024 Aug;10(8):1215-1230. doi: 10.1038/s41477-024-01746-4. Epub 2024 Jul 16. Nat Plants. 2024. PMID: 39014153 - Dynamic evolution of the heterochromatin sensing histone demethylase IBM1.
Zhang Y, Jang H, Luo Z, Dong Y, Xu Y, Kantamneni Y, Schmitz RJ. Zhang Y, et al. PLoS Genet. 2024 Jul 11;20(7):e1011358. doi: 10.1371/journal.pgen.1011358. eCollection 2024 Jul. PLoS Genet. 2024. PMID: 38991029 Free PMC article. - Integrated metabolomics and transcriptomics analysis highlight key pathways involved in the somatic embryogenesis of Darjeeling tea.
Awon VK, Dutta D, Banerjee S, Pal S, Gangopadhyay G. Awon VK, et al. BMC Genomics. 2024 Feb 23;25(1):207. doi: 10.1186/s12864-024-10119-2. BMC Genomics. 2024. PMID: 38395740 Free PMC article. - A cyclin D1 intrinsically disordered domain accesses modified histone motifs to govern gene transcription.
Jiao X, Di Sante G, Casimiro MC, Tantos A, Ashton AW, Li Z, Quach Y, Bhargava D, Di Rocco A, Pupo C, Crosariol M, Lazar T, Tompa P, Wang C, Yu Z, Zhang Z, Aldaaysi K, Vadlamudi R, Mann M, Skordalakes E, Kossenkov A, Du Y, Pestell RG. Jiao X, et al. Oncogenesis. 2024 Jan 8;13(1):4. doi: 10.1038/s41389-023-00502-1. Oncogenesis. 2024. PMID: 38191593 Free PMC article. - Binding by the Polycomb complex component BMI1 and H2A monoubiquitination shape local and long-range interactions in the Arabidopsis genome.
Yin X, Romero-Campero FJ, Yang M, Baile F, Cao Y, Shu J, Luo L, Wang D, Sun S, Yan P, Gong Z, Mo X, Qin G, Calonje M, Zhou Y. Yin X, et al. Plant Cell. 2023 Jun 26;35(7):2484-2503. doi: 10.1093/plcell/koad112. Plant Cell. 2023. PMID: 37070946 Free PMC article.
References
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 - PubMed
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 - PubMed
- Callebaut I, Courvalin JC, Mornon JP (1999) The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett 446: 189–193 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases