A comprehensive comparison of comparative RNA structure prediction approaches - PubMed (original) (raw)
Comparative Study
A comprehensive comparison of comparative RNA structure prediction approaches
Paul P Gardner et al. BMC Bioinformatics. 2004.
Abstract
Background: An increasing number of researchers have released novel RNA structure analysis and prediction algorithms for comparative approaches to structure prediction. Yet, independent benchmarking of these algorithms is rarely performed as is now common practice for protein-folding, gene-finding and multiple-sequence-alignment algorithms.
Results: Here we evaluate a number of RNA folding algorithms using reliable RNA data-sets and compare their relative performance.
Conclusions: We conclude that comparative data can enhance structure prediction but structure-prediction-algorithms vary widely in terms of both sensitivity and selectivity across different lengths and homologies. Furthermore, we outline some directions for future research.
Figures
Figure 1
RNA analysis. Current automated approaches to analysing homologous RNA sequences and structures usually follow one of three "plans". Plan A uses aligned sequences (usually produced by a standard multiple sequence alignment algorithm) to infer a consensus secondary structure from the evolutionary and energetic information contained in an alignment. This is a highly successful approach, but is limited to data-sets with sequence homology high enough for the alignment step to work yet divergent enough for detection of structurally consistent mutations. Plan B employs the "Sankoff algorithm" to simultaneously align and infer a consensus structure. This algorithm requires extreme amounts of memory and time. Plan C aligns RNA structures rather than sequences. This approach can be used in the rare situation where reliable structures are known. Representative algorithms which could be used for each plan are indicated within the figure.
Figure 2
Alignment consistency. A violation of RNA structural alignment consistency is shown (left), together with a possible correction (right) – see text for details. Note that the inconsistent alignment may maximise sequence similarity, showing 3 mismatches versus 1 mismatch and 2 indels, with the concrete outcome depending on the gap scoring used. Inconsistency is the reason why it is dangerous to align two structures in string representation by a standard sequence alignment algorithm. Inconsistency is hard to detect by human eye inspection, and structural alignments in databases are not always free from consistency violations.
Figure 3
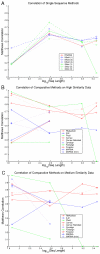
Prediction correlation with reality. Matthews correlation coefficient versus the logarithm of the sequence length for a range of different ncRNAs and structure prediction algorithms. Inset A shows accuracies of thermodynamic single sequence prediction algorithms. Insets B and C shows accuracies of comparative methods on the high and medium similarity data-sets respectively.
Figure 4
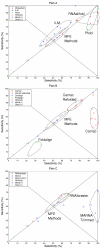
ROC plots. We use ROC (receiver operating characteristic) plots to simultaneously display both sensitivity and selectivity for plans A, B and C respectively. Accuracies of the MFE methods (MFold, RNAFold and SFold) are shown in each plot to provide a base-line. Points on the line X = Y are as sensitive as they are selective, points below this line indicates a greater selectivity, points above indicate greater sensitivity. Points below the line X = 100 - Y are worse than "random" assignments; Assuming base-pairs are independent of each other (this is false for base-pairing). Points in the top right corner are "perfect" predictions. Interestingly many algorithms form characteristic clusters in these plots. Where the variance is sufficiently small these have been indicated with a closed curve.
Similar articles
- Novel representation of RNA secondary structure used to improve prediction algorithms.
Zou Q, Lin C, Liu XY, Han YP, Li WB, Guo MZ. Zou Q, et al. Genet Mol Res. 2011 Sep 9;10(3):1986-98. doi: 10.4238/vol10-3gmr1181. Genet Mol Res. 2011. PMID: 21948761 - Secondary structure prediction for aligned RNA sequences.
Hofacker IL, Fekete M, Stadler PF. Hofacker IL, et al. J Mol Biol. 2002 Jun 21;319(5):1059-66. doi: 10.1016/S0022-2836(02)00308-X. J Mol Biol. 2002. PMID: 12079347 - Consensus shapes: an alternative to the Sankoff algorithm for RNA consensus structure prediction.
Reeder J, Giegerich R. Reeder J, et al. Bioinformatics. 2005 Sep 1;21(17):3516-23. doi: 10.1093/bioinformatics/bti577. Epub 2005 Jul 14. Bioinformatics. 2005. PMID: 16020472 - From consensus structure prediction to RNA gene finding.
Bernhart SH, Hofacker IL. Bernhart SH, et al. Brief Funct Genomic Proteomic. 2009 Nov;8(6):461-71. doi: 10.1093/bfgp/elp043. Brief Funct Genomic Proteomic. 2009. PMID: 19833701 Review. - Prediction and design of DNA and RNA structures.
Andersen ES. Andersen ES. N Biotechnol. 2010 Jul 31;27(3):184-93. doi: 10.1016/j.nbt.2010.02.012. Epub 2010 Mar 1. N Biotechnol. 2010. PMID: 20193785 Review.
Cited by
- CPU-GPU hybrid accelerating the Zuker algorithm for RNA secondary structure prediction applications.
Lei G, Dou Y, Wan W, Xia F, Li R, Ma M, Zou D. Lei G, et al. BMC Genomics. 2012;13 Suppl 1(Suppl 1):S14. doi: 10.1186/1471-2164-13-S1-S14. Epub 2012 Jan 17. BMC Genomics. 2012. PMID: 22369626 Free PMC article. - CoBold: a method for identifying different functional classes of transient RNA structure features that can impact RNA structure formation in vivo.
Martín AL, Mounir M, Meyer IM. Martín AL, et al. Nucleic Acids Res. 2021 Feb 26;49(4):e19. doi: 10.1093/nar/gkaa900. Nucleic Acids Res. 2021. PMID: 33095878 Free PMC article. - Predicting RNA secondary structure via adaptive deep recurrent neural networks with energy-based filter.
Lu W, Tang Y, Wu H, Huang H, Fu Q, Qiu J, Li H. Lu W, et al. BMC Bioinformatics. 2019 Dec 24;20(Suppl 25):684. doi: 10.1186/s12859-019-3258-7. BMC Bioinformatics. 2019. PMID: 31874602 Free PMC article. - GraphClust2: Annotation and discovery of structured RNAs with scalable and accessible integrative clustering.
Miladi M, Sokhoyan E, Houwaart T, Heyne S, Costa F, Grüning B, Backofen R. Miladi M, et al. Gigascience. 2019 Dec 1;8(12):giz150. doi: 10.1093/gigascience/giz150. Gigascience. 2019. PMID: 31808801 Free PMC article. - Efficient algorithms for probing the RNA mutation landscape.
Waldispühl J, Devadas S, Berger B, Clote P. Waldispühl J, et al. PLoS Comput Biol. 2008 Aug 8;4(8):e1000124. doi: 10.1371/journal.pcbi.1000124. PLoS Comput Biol. 2008. PMID: 18688270 Free PMC article.
References
- Poole AM, Jeffares DC, Penny D. The path from the RNA world. Journal of Molecular Evolution. 1998;46:1–17. - PubMed
- Jeffares DC, Poole AM, Penny D. Relics from the RNA world. Journal of Molecular Evolution. 1998;46:18–36. - PubMed
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, Tammana H, Gingeras TR. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Research. 2004;14:331–342. doi: 10.1101/gr.2094104. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases