Assigning African elephant DNA to geographic region of origin: applications to the ivory trade - PubMed (original) (raw)
Assigning African elephant DNA to geographic region of origin: applications to the ivory trade
Samuel K Wasser et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Resurgence of illicit trade in African elephant ivory is placing the elephant at renewed risk. Regulation of this trade could be vastly improved by the ability to verify the geographic origin of tusks. We address this need by developing a combined genetic and statistical method to determine the origin of poached ivory. Our statistical approach exploits a smoothing method to estimate geographic-specific allele frequencies over the entire African elephants' range for 16 microsatellite loci, using 315 tissue and 84 scat samples from forest (Loxodonta africana cyclotis) and savannah (Loxodonta africana africana) elephants at 28 locations. These geographic-specific allele frequency estimates are used to infer the geographic origin of DNA samples, such as could be obtained from tusks of unknown origin. We demonstrate that our method alleviates several problems associated with standard assignment methods in this context, and the absolute accuracy of our method is high. Continent-wide, 50% of samples were located within 500 km, and 80% within 932 km of their actual place of origin. Accuracy varied by region (median accuracies: West Africa, 135 km; Central Savannah, 286 km; Central Forest, 411 km; South, 535 km; and East, 697 km). In some cases, allele frequencies vary considerably over small geographic regions, making much finer discriminations possible and suggesting that resolution could be further improved by collection of samples from locations not represented in our study.
Figures
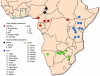
Fig. 1.
Map of Africa showing the collection sites divided into five regions: West Africa (cyan), Central forest (red), and Central (black), South (green), and East (blue) savannah.
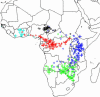
Fig. 2.
Estimated locations of elephant tissue and fecal samples from across Africa when assignments are allowed to vary anywhere within the elephants' range. All tissue and scat samples (n = 399) successfully amplified at seven or more loci. Sampling locations are indicated by a cross and are color-coded according to actual broad geographic region of origin: West Africa, Central forest, and Central, South, and East savannah (color-coded as in the Fig. 1 legend). Assigned location of each individual sample is shown by a circle and is color-coded according to its actual region of origin. The closer each circle is to crosses of the same color, the more accurate is that individual's assignment.
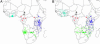
Fig. 3.
Representation of confidence of assignments when all neighboring samples from that subpopulation (represented by a single cross) were included (A) vs. excluded (B) from the calculation of geographic-specific allele frequencies for ET (green), Bia (cyan), and Mikumi (blue). The 100 color-coded circles are random draws from the set of all possible locations weighted according to their probability. The concentration of these 100 circles in any given area gives a guide to the probability that the sample arose from that area under each condition.
Similar articles
- Using DNA to track the origin of the largest ivory seizure since the 1989 trade ban.
Wasser SK, Mailand C, Booth R, Mutayoba B, Kisamo E, Clark B, Stephens M. Wasser SK, et al. Proc Natl Acad Sci U S A. 2007 Mar 6;104(10):4228-33. doi: 10.1073/pnas.0609714104. Epub 2007 Feb 26. Proc Natl Acad Sci U S A. 2007. PMID: 17360505 Free PMC article. - The evolution and phylogeography of the African elephant inferred from mitochondrial DNA sequence and nuclear microsatellite markers.
Eggert LS, Rasner CA, Woodruff DS. Eggert LS, et al. Proc Biol Sci. 2002 Oct 7;269(1504):1993-2006. doi: 10.1098/rspb.2002.2070. Proc Biol Sci. 2002. PMID: 12396498 Free PMC article. - The identification of elephant ivory evidences of illegal trade with mitochondrial cytochrome b gene and hypervariable D-loop region.
Lee EJ, Lee YH, Moon SH, Kim NY, Kim SH, Yang MS, Choi DH, Han MS. Lee EJ, et al. J Forensic Leg Med. 2013 Apr;20(3):174-8. doi: 10.1016/j.jflm.2012.06.014. Epub 2012 Jul 27. J Forensic Leg Med. 2013. PMID: 23472798 - Elephant natural history: a genomic perspective.
Roca AL, Ishida Y, Brandt AL, Benjamin NR, Zhao K, Georgiadis NJ. Roca AL, et al. Annu Rev Anim Biosci. 2015;3:139-67. doi: 10.1146/annurev-animal-022114-110838. Epub 2014 Dec 8. Annu Rev Anim Biosci. 2015. PMID: 25493538 Review. - Ivory Harvesting Pressure on the Genome of the African Elephant: A Phenotypic Shift to Tusklessness.
Raubenheimer EJ, Miniggio HD. Raubenheimer EJ, et al. Head Neck Pathol. 2016 Sep;10(3):332-5. doi: 10.1007/s12105-016-0704-y. Epub 2016 Feb 26. Head Neck Pathol. 2016. PMID: 26920555 Free PMC article. Review.
Cited by
- Genetic assignment of illegally trafficked neotropical primates and implications for reintroduction programs.
Oklander LI, Caputo M, Solari A, Corach D. Oklander LI, et al. Sci Rep. 2020 Feb 28;10(1):3676. doi: 10.1038/s41598-020-60569-3. Sci Rep. 2020. PMID: 32111905 Free PMC article. - Fast spatial ancestry via flexible allele frequency surfaces.
Rañola JM, Novembre J, Lange K. Rañola JM, et al. Bioinformatics. 2014 Oct 15;30(20):2915-22. doi: 10.1093/bioinformatics/btu418. Epub 2014 Jul 9. Bioinformatics. 2014. PMID: 25012181 Free PMC article. - Use of handheld X-ray fluorescence as a non-invasive method to distinguish between Asian and African elephant tusks.
Buddhachat K, Thitaram C, Brown JL, Klinhom S, Bansiddhi P, Penchart K, Ouitavon K, Sriaksorn K, Pa-in C, Kanchanasaka B, Somgird C, Nganvongpanit K. Buddhachat K, et al. Sci Rep. 2016 Apr 21;6:24845. doi: 10.1038/srep24845. Sci Rep. 2016. PMID: 27097717 Free PMC article. - Using DNA to track the origin of the largest ivory seizure since the 1989 trade ban.
Wasser SK, Mailand C, Booth R, Mutayoba B, Kisamo E, Clark B, Stephens M. Wasser SK, et al. Proc Natl Acad Sci U S A. 2007 Mar 6;104(10):4228-33. doi: 10.1073/pnas.0609714104. Epub 2007 Feb 26. Proc Natl Acad Sci U S A. 2007. PMID: 17360505 Free PMC article. - The use of DNA identification in prosecuting wildlife-traffickers in Australia: do the penalties fit the crimes?
Johnson RN. Johnson RN. Forensic Sci Med Pathol. 2010 Sep;6(3):211-6. doi: 10.1007/s12024-010-9174-9. Epub 2010 Jul 17. Forensic Sci Med Pathol. 2010. PMID: 20640536
References
- Dublin, H., Milliken, T. & Barnes, R. F. W. (1995) Four Years After the CITES Ban: Illegal Killings of Elephants, Ivory Trade and Stockpiles (International Union for Conservation of Nature and Natural Resources, Gland, Switzerland).
- Robinson, J. G. &Bennett, E. L. (1999) Hunting for Sustainability in Tropical Forests (Columbia Univ. Press, New York).
- Baker, C. S., Cipriano, F. & Paulmbi, S. R. (1996) Mol. Ecol. 5, 671–685.
- Douglas-Hamilton, I. (1987) Pachyderm 8, 1–10.
- Said, M. Y., Change, R. N., Craig, G. C., Thouless, C. R., Barnes, R. F. W. & Dublin, H. T. (1995) African Elephant Database (International Union for Conservation of Nature and Natural Resources, Gland, Switzerland).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources