CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis - PubMed (original) (raw)
CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis
Mark Leid et al. Gene Expr Patterns. 2004 Oct.
Abstract
Chicken ovalbumin upstream promoter transcription factor-interacting proteins 1 and 2 (CTIP1 and CTIP2) are related transcriptional regulatory proteins. While overexpression of both of these proteins has been linked to the development of several lymphoid malignancies, lack of CTIP1 and CTIP2 expression results in defective lymphopoiesis and abnormal thymocyte development, respectively. Here, we describe the expression patterns of CTIP1 and CTIP2 during mouse embryogenesis and in the post-natal brain. Both CTIP1 and CTIP2 were expressed diffusely in the embryo at 10.5 days post-coitum (d.p.c.). However, the expression of both genes became increasingly restricted to the central nervous system (CNS) during the course of fetal development, culminating with high, but differential, expression levels throughout the hippocampal subregions, olfactory bulb and cortex, limbic system, basal ganglia and frontal cortex of the developing brain, and in dorsal cells of the spinal cord. The brain expression domains of CTIP1 and CTIP2 were maintained into adulthood. Outside the CNS, both genes exhibited differential expression within the facial mesenchyme at 12.5 d.p.c., and CTIP2 was selectively expressed from day 12.5 onwards in the olfactory epithelium and developing thymus, and to a lesser extent in oral and gut epithelia. Strong CTIP2 expression was maintained in the thymus at 18.5 d.p.c. These results support the selective contributions of both CTIP1 and CTIP2 in the development and function of both the central nervous and immune systems and the importance of future investigations to define the function(s) of both proteins.
Figures
Fig. 1
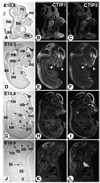
Expression of CTIP2 and CTIP2 during mouse development. Sagittal sections of mouse embryos or fetuses at 10.5 (A–C), 12.5 (D–F), 14.5 (G–I), and 18.5 d.p.c. (J–L), hybridized with antisense [35S]-labeled cRNAs corresponding to CTIP1 (middle column) and CTIP2 (right column), were examined by dark-field microscopy for signal detection (signal grain appears in white). A bright-field view of one of the corresponding sections is shown in the left column for histological orientation. ba, branchial arches; ce, cerebellum; fb, forebrain; flb, forelimb bud; gu, gut; hb, hindbrain; ht, heart; li, liver; mc, mouth cavity; nc, nasal cavity; ob, olfactory bulb; oe, esophagus; sc, spinal cord; st, striatum; te, telencephalic vesicle; th, thymus. Arrowheads in (E,F) point to differential spatial expression domains in the developing mandibular and frontonasal region.
Fig. 2
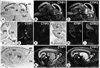
Expression of CTIP1 and CTIP2 in the developing mouse brain. Embryos were sagittally sectioned at 14.5 (A–F) and 18.5 d.p.c. (G–L), and hybridized with [35S]-labeled probes as indicated. ap, anterior pituitary; bg, basal ganglia; cc, cerebral cortex; ce, cerebellum; di, diencephalon; g5, 5th cranial nerve ganglion (trigeminal ganglion); hi, hippocampus; ht, hypothalamus; ic, inferior colliculus; me, mesencephalon; nc, nasal cavity; ob, olfactory bulb; po, pontine nucleus.
Fig. 3
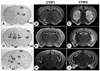
Mouse brain expression of CTIP1 and CTIP2 at post-natal day 21. Mouse brain coronal sections from post-natal day 21 animals were probed with [35S]-labeled riboprobes as indicated. A bright-field image is shown in the left column for histological orientation of the in situ hybridization panels (dark-field images in the middle and right columns). The sections shown are progressively more caudal from the top to the bottom rows. an, amydaloid nucleus; cc, cerebral cortex; cp, caudate-putamen; dg, dentate gyrus; hi, hippocampus; ht, hypothalamus, lateral septal nucleus; oc, olfactory cortex; pv, paraventricular thalamic nucleus; th, thalamus.
Fig. 4
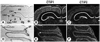
Expression of CTIP1 and CTIP2 in adult cerebral cortex, hippocampus, and cerebellum. Coronal cryosections of adult mouse brain were probed with [35S]-labeled CTIP1 (B) and CTIP2 (C) riboprobes. (A) Bright-field image for panels B and C. (D–F) Sagittal sections of adult mouse cerebellum hybridized with CTIP1 (E) and CTIP2 (F) probes and the corresponding bright-field image (D). CA1–CA4, hippocampal subregions; cc, cerebral cortex; hi, hippocampus; dg, dentate gyrus; th, thalamus; pl, Purkinje cell layer.
Similar articles
- Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression.
Senawong T, Peterson VJ, Avram D, Shepherd DM, Frye RA, Minucci S, Leid M. Senawong T, et al. J Biol Chem. 2003 Oct 31;278(44):43041-50. doi: 10.1074/jbc.M307477200. Epub 2003 Aug 19. J Biol Chem. 2003. PMID: 12930829 Free PMC article. - Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors.
Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. Avram D, et al. J Biol Chem. 2000 Apr 7;275(14):10315-22. doi: 10.1074/jbc.275.14.10315. J Biol Chem. 2000. PMID: 10744719 Free PMC article. - COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein.
Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M. Avram D, et al. Biochem J. 2002 Dec 1;368(Pt 2):555-63. doi: 10.1042/BJ20020496. Biochem J. 2002. PMID: 12196208 Free PMC article. - Expression of COUP-TF-interacting protein 2 (CTIP2) in mouse skin during development and in adulthood.
Golonzhka O, Leid M, Indra G, Indra AK. Golonzhka O, et al. Gene Expr Patterns. 2007 Aug;7(7):754-60. doi: 10.1016/j.modgep.2007.06.002. Epub 2007 Jun 13. Gene Expr Patterns. 2007. PMID: 17631058 Free PMC article. - CTIP2 and lipid metabolism: regulation in skin development and associated diseases.
Bhattacharya N, Ganguli-Indra G, Indra AK. Bhattacharya N, et al. Expert Rev Proteomics. 2021 Nov;18(11):1009-1017. doi: 10.1080/14789450.2021.2003707. Epub 2021 Nov 17. Expert Rev Proteomics. 2021. PMID: 34739354 Free PMC article. Review.
Cited by
- Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing.
Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, Aunis D, Rohr O. Marban C, et al. EMBO J. 2007 Jan 24;26(2):412-23. doi: 10.1038/sj.emboj.7601516. EMBO J. 2007. PMID: 17245431 Free PMC article. - Functional roles for the striatal-enriched transcription factor, Bcl11b, in the control of striatal gene expression and transcriptional dysregulation in Huntington's disease.
Desplats PA, Lambert JR, Thomas EA. Desplats PA, et al. Neurobiol Dis. 2008 Sep;31(3):298-308. doi: 10.1016/j.nbd.2008.05.005. Epub 2008 May 22. Neurobiol Dis. 2008. PMID: 18595722 Free PMC article. - Established monolayer differentiation of mouse embryonic stem cells generates heterogeneous neocortical-like neurons stalled at a stage equivalent to midcorticogenesis.
Sadegh C, Macklis JD. Sadegh C, et al. J Comp Neurol. 2014 Aug 15;522(12):2691-706. doi: 10.1002/cne.23576. Epub 2014 Apr 12. J Comp Neurol. 2014. PMID: 24610556 Free PMC article. - Kinetic analysis of BCL11B multisite phosphorylation-dephosphorylation and coupled sumoylation in primary thymocytes by multiple reaction monitoring mass spectroscopy.
Vogel WK, Gafken PR, Leid M, Filtz TM. Vogel WK, et al. J Proteome Res. 2014 Dec 5;13(12):5860-8. doi: 10.1021/pr5007697. Epub 2014 Nov 25. J Proteome Res. 2014. PMID: 25423098 Free PMC article. - CNS-specific regulatory elements in brain-derived HIV-1 strains affect responses to latency-reversing agents with implications for cure strategies.
Gray LR, Cowley D, Welsh C, Lu HK, Brew BJ, Lewin SR, Wesselingh SL, Gorry PR, Churchill MJ. Gray LR, et al. Mol Psychiatry. 2016 Apr;21(4):574-84. doi: 10.1038/mp.2015.111. Epub 2015 Aug 25. Mol Psychiatry. 2016. PMID: 26303660 Free PMC article.
References
- Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J. Biol. Chem. 2000;275:10315–10322. - PMC - PubMed
- Ghez C, Thack WT. The cerebellum. In: Kandel ER, et al., editors. Principles of Neural Science. New York: McGraw-Hill; 2000. pp. 832–852.
- Gunnersen JM, Augustine C, Spirkoska V, Kim M, Brown M, Tan SS. Global analysis of gene expression patterns in developing mouse neocortex using serial analysis of gene expression. Mol. Cell. Neurosci. 2002;19:560–573. - PubMed
- Jonk LJ, de Jonge ME, Pals CE, Wissink S, Vervaart JM, Schoorlemmer J, Kruijer W. Cloning and expression during development of three murine members of the COUP family of nuclear orphan receptors. Mech. Dev. 1994;47:81–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 ES000210-35/ES/NIEHS NIH HHS/United States
- ES00210/ES/NIEHS NIH HHS/United States
- GM60852/GM/NIGMS NIH HHS/United States
- R01 GM060852/GM/NIGMS NIH HHS/United States
- R01 GM060852-02/GM/NIGMS NIH HHS/United States
- P30 ES000210/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases