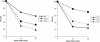Effects of the U1C L13 mutation and temperature regulation of yeast commitment complex formation - PubMed (original) (raw)
Effects of the U1C L13 mutation and temperature regulation of yeast commitment complex formation
Hansen Du et al. Proc Natl Acad Sci U S A. 2004.
Abstract
The U1 small nuclear ribonucleoprotein particle U1C protein has a zinc finger-like structure (C2H2 motif) at its N terminus, which is conserved from yeast to humans. Mutations of amino acid L13 within this domain rescue the essential function of the helicase protein Prp28p. Prp28p has been implicated in unwinding the 5' splice site (5'ss)-U1 small nuclear RNA (snRNA) base-pairing, to allow replacement of U1 snRNA with U6 snRNA during spliceosome assembly. The L13 phenotype has therefore been interpreted to indicate that WT U1C contributes to 5'ss-U1 snRNA stabilization by binding to the RNA duplex. We show here that an L13 mutant extract cannot form stable base-pairing at room temperature but is permissive for U1-5'ss base-pairing at low temperature. This phenotype is similar to that of a U1C-depleted extract, indicating that the U1C L13 mutation is a strong loss-of-function mutation. The two mutant extracts are unlike a WT extract, which undergoes stable pairing at room temperature but little or no pairing at low temperature. Taken together with previous results and the failure to observe a direct interaction of U1C with the U1-5'ss duplex, the data suggest that U1C contributes indirectly to stable U1-5'ss base-pairing under permissive conditions. A model is proposed to account for the L13 results.
Figures
Fig. 1.
The effect of U1C on U1 snRNA–5′ss base-pairing at normal and low temperatures. (A) RNA–RNA base-pairing assayed by psoralen cross-linking at both high and low temperatures. Standard commitment complex formation assays were performed in WT, U1C L13 mutant, and U1C depletion extracts with 32P-radiolabeled WT-72 at the two temperatures. Psoralen cross-linking was carried out after 20 min of incubation as described (20). U1 snRNA–5′ss interactions and free RNA substrates are indicated by arrows. (B) Percentage of low-temperature base-pairing. The relative intensity of lanes 4–6 of A was measured, and the base-pairing in the U1C-depleted extract at low temperature was arbitrarily set to 100%.
Fig. 2.
Stability of the U1 snRNA–5′ss base-pairing at two different temperatures. Chase assays were used to measure the stability of RNA–RNA interactions formed in the three different extracts. 32P-labeled transcripts were incubated with splicing extracts in standard commitment complex reactions for 20 min at either 25°C (A) or 0°C (B). Aliquots were removed, and a 400-fold molar excess of unlabeled RNAs was added. Psoralen was then added at the times indicated. The samples were UV-irradiated as described (20).
Fig. 3.
Stability of the RNA–protein complexes formed in three different extracts at 25°C (Left) and 0°C (Right). Commitment complex formation and chase assays were carried out as described in Materials and Methods. Complex stability was measured by liquid scintillation counter after immunoprecipitation with an antibody against Prp40. The value obtained after a 20-min incubation without unlabeled RNA was arbitrarily set to 100. Values were averaged from at least four experiments.
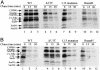
Fig. 4.
Stability comparisons of RNA–protein interactions in the three extracts at two different incubation temperatures: 25°C(A) and 0°C(B). 4-Thio-UTP-labeled 32P WT-72 RNA (9) was used as a substrate for protein UV cross-linking. Results for the WT and RNase-H-treated extract at 25°C were as described (9, 20, 25). Previously identified proteins are indicated on the left.
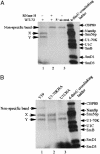
Fig. 5.
MB-mediated cross-linking to detect protein–double-stranded RNA interactions. (A) WT extract. (B) Comparison of WT and tagged extracts.32P-labeled transcripts were incubated with splicing extracts in standard commitment complex reactions for 20 min at 25°C. MB cross-linking was then carried out as described in Materials and Methods. Protein size ladder was the profile of WT-72 4-thio-UTP UV cross-linking. The intensity of a large nonspecific band was increased when that of the two specific bands was decreased (compare lane 1 with lanes 2 and 3 in A).
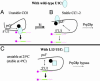
Fig. 6.
Working model for the role of yeast U1C during in vitro commitment complex formation. Protein–protein interactions contribute to retaining WT U1C (blue oval, A and B; blue square, C) within U1 snRNP. The black ball is a putative U1 snRNP protein, which helps retain U1C within the snRNP when U1C interactions with U1 snRNA are weak or negligible (e.g., in A because of an RNA conformational change or in C because of a U1C protein conformational change). The colored bar is the pre-mRNA, with the intron in green. (A) There is little U1C binding to U1 RNA at low temperature, because of a favored RNA–RNA interaction, depicted as an intramolecular base-pairing interaction between a putative ps5′ and the 5′ss. Under these conditions, the 5′ substrate interaction is of low stability (indicated by a thick arrow in A) and predominantly with U1 snRNP proteins including U1C. (B) At higher temperatures (indicated by kt), there is a conformational change (indicated by a double arrow in A) that includes U1C binding to the ps5′. This process liberates the 5′ss and allows a base-pairing interaction with the pre-mRNA 5′ss (indicated by thin lines), which gives rise to a more stable interaction. (C) The L13 mutation causes a U1C conformational change (blue square rather than blue oval), which binds poorly to RNA. U1 RNA therefore has a tendency to adopt the low-temperature conformation as in A, which allows the ps5′ to interact with the 5′ end of U1 (double arrow) and compete with pre-mRNA base-pairing. This process gives rise to a lower stability base-pairing interaction (thick arrow).
Similar articles
- The U1 snRNP protein U1C recognizes the 5' splice site in the absence of base pairing.
Du H, Rosbash M. Du H, et al. Nature. 2002 Sep 5;419(6902):86-90. doi: 10.1038/nature00947. Nature. 2002. PMID: 12214237 - Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5'ss base pairing in yeast.
Lacadie SA, Rosbash M. Lacadie SA, et al. Mol Cell. 2005 Jul 1;19(1):65-75. doi: 10.1016/j.molcel.2005.05.006. Mol Cell. 2005. PMID: 15989965 - A targeted bypass screen identifies Ynl187p, Prp42p, Snu71p, and Cbp80p for stable U1 snRNP/Pre-mRNA interaction.
Hage R, Tung L, Du H, Stands L, Rosbash M, Chang TH. Hage R, et al. Mol Cell Biol. 2009 Jul;29(14):3941-52. doi: 10.1128/MCB.00384-09. Epub 2009 May 18. Mol Cell Biol. 2009. PMID: 19451230 Free PMC article. - Principles and correction of 5'-splice site selection.
Malard F, Mackereth CD, Campagne S. Malard F, et al. RNA Biol. 2022 Jan;19(1):943-960. doi: 10.1080/15476286.2022.2100971. RNA Biol. 2022. PMID: 35866748 Free PMC article. Review. - Recognition of the 5' splice site by the spliceosome.
Konarska MM. Konarska MM. Acta Biochim Pol. 1998;45(4):869-81. Acta Biochim Pol. 1998. PMID: 10397335 Review.
Cited by
- Differentially expressed, variant U1 snRNAs regulate gene expression in human cells.
O'Reilly D, Dienstbier M, Cowley SA, Vazquez P, Drozdz M, Taylor S, James WS, Murphy S. O'Reilly D, et al. Genome Res. 2013 Feb;23(2):281-91. doi: 10.1101/gr.142968.112. Epub 2012 Oct 15. Genome Res. 2013. PMID: 23070852 Free PMC article. - The U11-48K protein contacts the 5' splice site of U12-type introns and the U11-59K protein.
Turunen JJ, Will CL, Grote M, Lührmann R, Frilander MJ. Turunen JJ, et al. Mol Cell Biol. 2008 May;28(10):3548-60. doi: 10.1128/MCB.01928-07. Epub 2008 Mar 17. Mol Cell Biol. 2008. PMID: 18347052 Free PMC article. - Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing.
Selvanathan SP, Graham GT, Erkizan HV, Dirksen U, Natarajan TG, Dakic A, Yu S, Liu X, Paulsen MT, Ljungman ME, Wu CH, Lawlor ER, Üren A, Toretsky JA. Selvanathan SP, et al. Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):E1307-16. doi: 10.1073/pnas.1500536112. Epub 2015 Mar 3. Proc Natl Acad Sci U S A. 2015. PMID: 25737553 Free PMC article. - DExD/H-box Prp5 protein is in the spliceosome during most of the splicing cycle.
Kosowski TR, Keys HR, Quan TK, Ruby SW. Kosowski TR, et al. RNA. 2009 Jul;15(7):1345-62. doi: 10.1261/rna.1065209. Epub 2009 May 18. RNA. 2009. PMID: 19451545 Free PMC article. - Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases.
Jankowsky E, Bowers H. Jankowsky E, et al. Nucleic Acids Res. 2006;34(15):4181-8. doi: 10.1093/nar/gkl410. Epub 2006 Aug 25. Nucleic Acids Res. 2006. PMID: 16935886 Free PMC article. Review.
References
- Moore, M. J., Query, C. C. & Sharp, P. A. (1993) in The RNA World, eds. Gesteland, R. F. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 303–357.
- Will, C. L. & Luhrmann, R. (1997) Curr. Opin. Cell Biol. 9, 320–328. - PubMed
- Burge, C. B., Tuschl, T. & Sharp, P. A. (1999) in The RNA World II, eds. Gesteland, R. R., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 525–560.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases