Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi - PubMed (original) (raw)
Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi
Jennifer M Henke et al. J Bacteriol. 2004 Oct.
Abstract
In a process called quorum sensing, bacteria communicate using extracellular signal molecules termed autoinducers. Two parallel quorum-sensing systems have been identified in the marine bacterium Vibrio harveyi. System 1 consists of the LuxM-dependent autoinducer HAI-1 and the HAI-1 sensor, LuxN. System 2 consists of the LuxS-dependent autoinducer AI-2 and the AI-2 detector, LuxPQ. The related bacterium, Vibrio cholerae, a human pathogen, possesses System 2 (LuxS, AI-2, and LuxPQ) but does not have obvious homologues of V. harveyi System 1. Rather, System 1 of V. cholerae is made up of the CqsA-dependent autoinducer CAI-1 and a sensor called CqsS. Using a V. cholerae CAI-1 reporter strain we show that many other marine bacteria, including V. harveyi, produce CAI-1 activity. Genetic analysis of V. harveyi reveals cqsA and cqsS, and phenotypic analysis of V. harveyi cqsA and cqsS mutants shows that these functions comprise a third V. harveyi quorum-sensing system that acts in parallel to Systems 1 and 2. Together these communication systems act as a three-way coincidence detector in the regulation of a variety of genes, including those responsible for bioluminescence, type III secretion, and metalloprotease production.
Figures
FIG. 1.
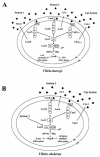
Models of the V. harveyi and V. cholerae quorum-sensing systems. (A) V. harveyi has three parallel quorum-sensing systems that regulate the genes encoding bioluminescence (Lux), TTS, a secreted metalloprotease (VhpA), and other quorum-sensing-activated and -repressed genes. (B) V. cholerae possesses the CAI-1-CqsS system and the AI-2-LuxPQ system. V. cholerae also has an unidentified third circuit (System 3) that acts through LuxO. Symbols: pentagons, HAI-1; diamonds, AI-2; circles, CAI-1. Dotted lines denote targets for which regulation has not been shown to be direct. In both panels, phosphate flows toward LuxO at low cell density, and the flow of phosphate is reversed at high cell density (double-headed arrows). Details of the phosphorelay mechanism are provided in the text. Abbreviations: H, histidine; D, aspartate; HTH, helix-turn-helix; IM, inner membrane; OM, outer membrane.
FIG. 2.
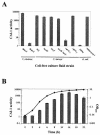
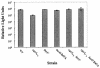
Analysis of cell-free culture fluids for CAI-1 activity. (A) Cell-free culture fluids were prepared from the strains shown and added at 30% (vol/vol) to V. cholerae MM920. Light production by MM920 was measured. Strain names are as follows (left to right): KSK1052, MM893, BB120, JAF633, KM387, KM413, JMH603, JMH598, DH5α, MM1467. (B) CAI-1 production over the growth curve is shown for WT V. harveyi strain BB120. Samples were collected every 2 h, the OD600s were measured, and cell-free culture fluids were prepared and assayed for CAI-1 activity using MM920. CAI-1 activity is reported as the fold induction of light production in MM920.
FIG. 3.
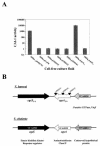
The cqs locus in V. harveyi. (A) CAI-1 activity is shown for recombinant E. coli DH5α strains carrying either the cosmid pLAFR or pLAFR containing vibrio DNA. pJMH17F7 carries 25 kb of V. harveyi genomic DNA containing the gene responsible for CAI-1 activity. pJMH159, pJMH163, and pJMH164 contain Tn_5lacZ_ insertions in pJMH17F7. pJMH273 carries only the cqsAVh gene, and pJMH282 carries only the cqsSVh gene. (B) Map of the cqs locus of V. harveyi and V. cholerae. Black triangles indicate the sites of Tn_5lacZ_ insertions in pJMH163, pJMH164 and pJMH159. The cqsS and cqsA genes are shown in black and white, respectively. The downstream ORF in V. harveyi has amino acid homology to VCA0528 and is shown in gray. This gene was not fully sequenced. The downstream ORF in V. cholerae, denoted VCA0524, is shown in stripes.
FIG. 4.
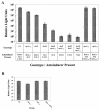
V. harveyi single, double, and triple autoinducer synthase mutant phenotypes. (A) Light production per cell (RLU) was measured from high-cell-density V. harveyi cultures. The V. harveyi strains are (from left to right) BB120 (WT), JMH603 (cqsAVh), KM387 (luxS), JAF633 (luxM), JMH605 (luxM cqsAVh), JMH606 (luxS cqsAVh), KM413 (luxM luxS), and JMH634 (luxM luxS cqsAVh), and the rightmost two bars are JMH603 (cqsAVh). In these latter two cases, JMH603 (cqsAVh) was grown in the presence of 30% V. harveyi or V. cholerae WT culture fluids. The asterisks denote exogenously added autoinducer. (B) CqsA_Vh_ and CqsS_Vh_ epistasis analysis. RLU were measured at high cell density in the following strains (left to right): BB120 (WT), JMH603 (cqsAVh), JMH598 (cqsSVh), JMH763 (cqsAVh cqsSVh). Relative light units are defined as counts min−1 ml−1 × 1,000/CFU ml−1.
FIG. 5.
LuxU and LuxO are epistatic to CqsA. RLU were measured from high-cell-density V. harveyi cultures of the following strains (left to right): BB120 (WT), JMH603 (cqsAVh), JAF536 (luxU), JAF483 (_luxO_D47A), JMH621 (luxU cqsAVh), JMH774 (_luxO_D47A cqsAVh).
FIG. 6.
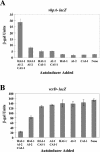
All three autoinducers are required for quorum-sensing-controlled gene regulation. β-Galactosidase assays were performed on V. harveyi recipient strains (high cell density) grown overnight in the presence of 10% cell-free culture fluids containing various autoinducers. Cell-free culture fluids were prepared from the following strains (left to right): BB120 (WT), JMH603 (cqsAVh), KM387 (luxS), JAF633 (luxM), JMH606 (luxS cqsAVh), JMH605 (luxM cqsAVh), KM413 (luxM luxS), and JMH634 (luxM luxS cqsAVh). (A) The recipient strain was V. harveyi JMH670 (vhpA::Tn_5lacZ luxM luxS cqsAVh_). (B) The recipient strain was JMH674 (vcrD::Tn_5lacZ luxM luxS cqsAVh_). β-Galactosidase units are reported as (Vmax × dilution factor)/OD600.
FIG. 7.
V. harveyi single, double, and triple autoinducer sensor mutant phenotypes. Density-dependent bioluminescence assays were performed on (A) BB120 (WT), BB721 (luxO), BB170 (luxN), BB886 (luxQ), and JMH598 (cqsSVh); (B) BB120 (WT), BB721 (luxO), JMH612 (luxQ cqsSVh), JMH597 (luxN cqsSVh), JMH625 (luxN luxQ), and JMH626 (luxN luxQ cqsAVh); and (C) BB120 (WT), BB721 (luxO), JMH625 (luxN luxQ), and JMH628 (luxN luxQ cqsSVh). Relative light units are reported as counts min−1 ml−1 × 1,000/CFU ml−1. In panels A and B, cells were diluted 1:5,000, and in panel C, cells were diluted 1:500,000 prior to the start of the experiment.
Similar articles
- Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression.
Mok KC, Wingreen NS, Bassler BL. Mok KC, et al. EMBO J. 2003 Feb 17;22(4):870-81. doi: 10.1093/emboj/cdg085. EMBO J. 2003. PMID: 12574123 Free PMC article. - Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus.
Henke JM, Bassler BL. Henke JM, et al. J Bacteriol. 2004 Jun;186(12):3794-805. doi: 10.1128/JB.186.12.3794-3805.2004. J Bacteriol. 2004. PMID: 15175293 Free PMC article. - Quorum-sensing autoinducer molecules produced by members of a multispecies biofilm promote horizontal gene transfer to Vibrio cholerae.
Antonova ES, Hammer BK. Antonova ES, et al. FEMS Microbiol Lett. 2011 Sep;322(1):68-76. doi: 10.1111/j.1574-6968.2011.02328.x. Epub 2011 Jun 27. FEMS Microbiol Lett. 2011. PMID: 21658103 - Oxazaborolidine derivatives inducing autoinducer-2 signal transduction in Vibrio harveyi.
Aharoni R, Bronstheyn M, Jabbour A, Zaks B, Srebnik M, Steinberg D. Aharoni R, et al. Bioorg Med Chem. 2008 Feb 15;16(4):1596-604. doi: 10.1016/j.bmc.2007.11.032. Epub 2007 Nov 17. Bioorg Med Chem. 2008. PMID: 18053731 Review. - LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria.
Winzer K, Hardie KR, Williams P. Winzer K, et al. Adv Appl Microbiol. 2003;53:291-396. doi: 10.1016/s0065-2164(03)53009-x. Adv Appl Microbiol. 2003. PMID: 14696323 Review. No abstract available.
Cited by
- A Vibrio cholerae Type IV restriction system targets glucosylated 5-hydroxymethylcytosine to protect against phage infection.
Gomez JB, Waters CM. Gomez JB, et al. J Bacteriol. 2024 Sep 19;206(9):e0014324. doi: 10.1128/jb.00143-24. Epub 2024 Sep 4. J Bacteriol. 2024. PMID: 39230524 - Structural and functional insights underlying recognition of histidine phosphotransfer protein in fungal phosphorelay systems.
Paredes-Martínez F, Eixerés L, Zamora-Caballero S, Casino P. Paredes-Martínez F, et al. Commun Biol. 2024 Jul 4;7(1):814. doi: 10.1038/s42003-024-06459-0. Commun Biol. 2024. PMID: 38965424 Free PMC article. - Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Zhang J, Liu B, Gu D, Hao Y, Chen M, Ma Y, Zhou X, Reverter D, Zhang Y, Wang Q. Zhang J, et al. Nucleic Acids Res. 2021 Apr 6;49(6):3274-3293. doi: 10.1093/nar/gkab150. Nucleic Acids Res. 2021. PMID: 33693882 Free PMC article. - Requirements for Vibrio cholerae HapR binding and transcriptional repression at the hapR promoter are distinct from those at the aphA promoter.
Lin W, Kovacikova G, Skorupski K. Lin W, et al. J Bacteriol. 2005 May;187(9):3013-9. doi: 10.1128/JB.187.9.3013-3019.2005. J Bacteriol. 2005. PMID: 15838027 Free PMC article. - Probing bacterial transmembrane histidine kinase receptor-ligand interactions with natural and synthetic molecules.
Ng WL, Wei Y, Perez LJ, Cong J, Long T, Koch M, Semmelhack MF, Wingreen NS, Bassler BL. Ng WL, et al. Proc Natl Acad Sci U S A. 2010 Mar 23;107(12):5575-80. doi: 10.1073/pnas.1001392107. Epub 2010 Mar 8. Proc Natl Acad Sci U S A. 2010. PMID: 20212168 Free PMC article.
References
- Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. - PubMed
- Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. - PubMed
- Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol. 12:403-412. - PubMed
- Beringer, J. E., J. L. Beynon, A. V. Buchanan-Wollaston, and A. W. B. Johnston. 1978. Transfer of the drug resistance transposon Tn5 to Rhizobium. Nature 276:633-634.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous