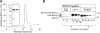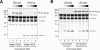Sec1p directly stimulates SNARE-mediated membrane fusion in vitro - PubMed (original) (raw)
Sec1p directly stimulates SNARE-mediated membrane fusion in vitro
Brenton L Scott et al. J Cell Biol. 2004.
Abstract
Sec1 proteins are critical players in membrane trafficking, yet their precise role remains unknown. We have examined the role of Sec1p in the regulation of post-Golgi secretion in Saccharomyces cerevisiae. Indirect immunofluorescence shows that endogenous Sec1p is found primarily at the bud neck in newly budded cells and in patches broadly distributed within the plasma membrane in unbudded cells. Recombinant Sec1p binds strongly to the t-SNARE complex (Sso1p/Sec9c) as well as to the fully assembled ternary SNARE complex (Sso1p/Sec9c;Snc2p), but also binds weakly to free Sso1p. We used recombinant Sec1p to test Sec1p function using a well-characterized SNARE-mediated membrane fusion assay. The addition of Sec1p to a traditional in vitro fusion assay moderately stimulates fusion; however, when Sec1p is allowed to bind to SNAREs before reconstitution, significantly more Sec1p binding is detected and fusion is stimulated in a concentration-dependent manner. These data strongly argue that Sec1p directly stimulates SNARE-mediated membrane fusion.
Figures
Figure 1.
Production of Sec1p and characterization of Sec1p antisera. (A) Recombinant His6-Sec1p production. Size exclusion chromatogram. His6-Sec1p migrates as a single species and elutes slightly slower than BSA (67,000 D, middle arrowhead) on a Superose 12 (HR 10/30) column. Left arrowhead is 200,000 D (β-amylase) and the right arrowhead is 12,400 D (Cytochrome c). (Inset) Coomassie blue–stained gel of purified Sec1p. Lane 1 is a pooled fraction from metal chelate chromatography. This pool was further purified by ion-exchange chromatography, shown in lane 2. (B) Sec1p antibody production and yeast overexpression. The specificity of the polyclonal antisera is shown by detection of endogenous Sec1p in cytosolic extracts of the BY4741pep4Δ strain carrying the empty parent vector pYX223, and the detection of overexpressed Sec1p (2Xmyc-His6-Sec1p, pJM255). The degree of overproduction is also measured with the Sec1p antisera in comparison to a dilution series of recombinant Sec1p from E. coli.
Figure 2.
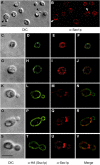
Immunolocalization of endogenous Sec1p. Sec1p localizes to patches on the plasma membrane. (A) Differential interference contrast (DIC) image of S. cerevisiae. (B) Endogenous Sec1p is imaged in the field of cells shown in A using a polyclonal anti-Sec1p antibody. Arrowheads denote newly emerged buds. (C–V) Individual cells in different stages of the cell cycle are imaged: Small, unbudded cells (C–J), small-budded cells (K–R), and a large-budded cell (S–V). DIC images (C, G, K, O, and S) and indirect immunofluorescence images are shown for each cell. Sso1p localization was determined by staining a HA-tagged Sso1p with anti-HA (D, H, L, P, and T). Endogenous Sec1p localization in individual cells is illustrated (E, I, M, Q, and U) and a merge of both Sso1p and Sec1p staining is imaged (F, J, N, R, and V). We determined that 71 ± 15% of endogenous Sec1p colocalizes with Sso1p-HA (n = 62 cells) when total cell area is examined. Bars, 5 μm.
Figure 3.
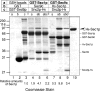
Sec1p binds strongly to the t-SNARE complex (Sso1p/Sec9c) and the fully assembled ternary SNARE complex (Sso1p/Sec9c;Snc2p). GST pull-down experiments were used to assess the degree of Sec1p binding to various SNAREs and SNARE complexes. The level of nonspecific binding was determined by incubation of recombinant Sec1p with protein-free GSH beads (lane 1), GST (lane 2), or GST-Sed5p (lane 3). The level of Sec1p binding to GST-Sed5p (which was approximately four times higher than either protein free beads or GST alone) was used as the background value for quantitative analysis. Sec1p binding to three SNARE species was analyzed: free t-SNARE (lanes 4 and 7), the binary t-SNARE complex (lanes 5 and 8) or the ternary SNARE complex (lanes 6 and 9). The three species were attached to glutathione beads via GST-Sso1p (lanes 4–6) or GST-Sec9c (lanes 7–9). Purified recombinant Sec1p (lane 10) is also shown (∼1.4 μg, ∼16 pmol). Sec1p binding was quantified by densitometry and is represented as fold above the GST-Sed5p background. Sec1p binds most significantly to the t-SNARE complex (lanes 5 and 8) as well as the ternary SNARE complex (lanes 6 and 9). A weaker association was also seen with the free monomeric SNAREs GST-Sso1p (lane 4). No binding to free GST-Sec9c (lane 7) was detected above background.
Figure 4.
Effect of Sec1p on SNARE complex formation. (A) Binary t-SNARE complex and ternary SNARE complex formation is unaffected by Sec1p when all components are added simultaneously. GST-Sec9c was bound to resin and equimolar amounts of His8-Sso1p, Snc2p-His6, and increasing amounts of His6-Sec1p (0, 0.1, 0.2, and 0.4 nmol) were added at the same time and incubated at 4°C for ∼16 h. Bound complexes were resolved by SDS-PAGE and stained with Coomassie blue. (B) Snc2p can efficiently bind to Sec1p bound t-SNARE complexes without significant displacement of Sec1p. Increasing amounts of Snc2p (0, lane 1 and 5, 0.4 nmol lane 2 and 6, 0.8 nmol, lanes 3 and 7 and 1.6 nmol, lanes 4 and 8) were allowed to associate with preformed t-SNARE complex (Sso1p/Sec9c, lanes 1–4) or Sec1p bound t-SNARE complexes (Sec1p:Sso1p/Sec9c, lanes 5–8) for ∼16 h at 4°C. Bound complexes were resolved by SDS-PAGE and stained with Coomassie blue.
Figure 5.
Sec1p stimulates in vitro fusion. Recombinant Sec1p was added to an in vitro fusion assay containing reduced levels of SNARE proteins. 10 μl of t-SNARE liposomes (Sso1p/Sec9c, ∼19.5 μg, ∼215 pmol of t-SNARE complex proteins, ∼22.5 nmol lipid) was mixed with 85 μl of recombinant His6-Sec1p (∼12.8 μg, ∼150 pmol, closed circles) or buffer A200 (open circles) for ∼15 h at 4°C. 5 μl of Snc1p liposomes (∼8.3 μg, 630 pmol Snc1p and 1.95 nmol lipid) were added and the NBD fluorescence measured for 2 h in a fluorescent plate reader at 37°C. The background values (solid and dashed lines) represent an inhibited reaction containing the same components as stimulated fusion reaction in addition to the soluble domain of Snc2p to inhibit vesicle fusion. The amount of fusion at 120 min was 0.98 rounds of fusion for the Sec1p stimulated curve (closed circles), 0.74 rounds of fusion for basal fusion (open circles), compared with an inhibited background of 0.05 rounds of fusion. This experiment was repeated three additional times using independent recombinant Sec1p purifications. The average stimulation observed of the four experiments was 39.3% ± an SEM of 3.3%. In addition, each sample was fused with protein free fluorescently labeled liposomes that showed a background fusion level of 0.059 rounds of fusion (not depicted).
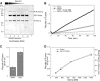
Figure 6.
SNARE-bound Sec1p strongly stimulates in vitro fusion. A twofold dilution series of Sec1p was bound to t-SNARE complexes (Sso1p/Sec9c) in detergent solution before vesicle reconstitution. (A) Coomassie blue–stained gel of liposomes containing Sec1p bound t-SNARE complexes. Acceptor t-SNARE liposomes containing various amounts of bound His6-Sec1p were resolved on a 10% Bis-Tris NuPAGE gel (Invitrogen) and stained with Coomassie blue. Lane 1 contains 15 μl of liposomes derived from a reaction containing His8-Sso1p (∼160 μg, ∼4.7 nmol) and 60 μg (700 pmol) of His6-Sec1p. Lanes 2–6 contains 10 μl of liposomes derived from reactions including His8-Sso1p (∼160 μg, ∼4.7 nmol), GST-Sec9c (∼430 μg, ∼7.7 nmol) and decreasing amounts of Sec1p: lane 2, ∼60, μg, ∼700 pmol; lane 3, ∼30 μg, ∼350 pmol; lane 4, ∼15 μg, ∼175 pmol; and lane 5, ∼7.5 μg, ∼88 pmol. Lane 6 contained no Sec1p. Lane 7 contains 0.6 μg (∼7 pmol) of recombinant His6-Sec1p. (B) Kinetic fusion graph of Sec1p stimulated fusion. Vesicles (45 μl) containing t-SNARE complexes without Sec1p (open circles, ∼13 μg, 145 pmol Sso1p/Sec9c and 42 nmol lipid) and t-SNARE vesicles containing the highest amount of bound Sec1p (closed circles) were mixed with fluorescently labeled vesicles containing the v-SNARE Snc1p (5 μl, ∼8.3 μg, 630 pmol Snc1p and 1.95 nmol lipid) and incubated for 120 min at 37°C in a standard fusion reaction. The extent of fusion is represented as rounds of fusion, measured as fold lipid dilution in the reaction. The background values (solid and dashed lines) represent an inhibited reaction containing the same components in addition to the soluble domain of Snc2p to inhibit vesicle fusion. The amount of fusion at 120 min was 1.32 rounds of fusion for the Sec1p stimulated curve (closed circles), 0.42 rounds of fusion for basal fusion (buffer, open circles), and the inhibited fusion background, 0.1 rounds of fusion. Sec1p stimulated fusion ∼3.8-fold in this experiment after background subtraction. (C) Average fold stimulation caused by Sec1p. The amount of stimulation by Sec1p was examined using four independent preparations of recombinant His6-Sec1p. This histogram shows that His6-Sec1p stimulates fusion by 2.7-fold on average. The mean ± SEM are represented after the subtraction of an average background of 0.105 rounds of fusion. (D) Sec1p titration showing stimulation is concentration dependent. The extent of fusion observed at 120 min and the amount of Sec1p binding detected to t-SNARE liposomes is represented for independent in vitro fusion experiments relative to the amount of Sec1p added to the reaction. Rounds of fusion at 120 min are shown on the left y axis (open circles) and the amount of Sec1p binding (relative to His8-Sso1p) is quantified on the right y axis (closed circles). Both values are plotted relative to the concentration of Sec1p added to the reaction (nM). The binding values for Sec1p were determined by quantifying the gel shown in Fig. 6 A.
Similar articles
- Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity.
Parlati F, Varlamov O, Paz K, McNew JA, Hurtado D, Söllner TH, Rothman JE. Parlati F, et al. Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5424-9. doi: 10.1073/pnas.082100899. Proc Natl Acad Sci U S A. 2002. PMID: 11959998 Free PMC article. - Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p.
Nicholson KL, Munson M, Miller RB, Filip TJ, Fairman R, Hughson FM. Nicholson KL, et al. Nat Struct Biol. 1998 Sep;5(9):793-802. doi: 10.1038/1834. Nat Struct Biol. 1998. PMID: 9731774 - Compartmental specificity of cellular membrane fusion encoded in SNARE proteins.
McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, Söllner TH, Rothman JE. McNew JA, et al. Nature. 2000 Sep 14;407(6801):153-9. doi: 10.1038/35025000. Nature. 2000. PMID: 11001046 - The role of Sec1p-related proteins in vesicle trafficking in the nerve terminal.
Pevsner J. Pevsner J. J Neurosci Res. 1996 Jul 15;45(2):89-95. doi: 10.1002/(SICI)1097-4547(19960715)45:2<89::AID-JNR1>3.0.CO;2-B. J Neurosci Res. 1996. PMID: 8843026 Review. - For better or for worse: complexins regulate SNARE function and vesicle fusion.
Brose N. Brose N. Traffic. 2008 Sep;9(9):1403-13. doi: 10.1111/j.1600-0854.2008.00758.x. Epub 2008 Apr 28. Traffic. 2008. PMID: 18445121 Review.
Cited by
- Functional Roles of UNC-13/Munc13 and UNC-18/Munc18 in Neurotransmission.
Meunier FA, Hu Z. Meunier FA, et al. Adv Neurobiol. 2023;33:203-231. doi: 10.1007/978-3-031-34229-5_8. Adv Neurobiol. 2023. PMID: 37615868 Review. - Base-edited cynomolgus monkeys mimic core symptoms of STXBP1 encephalopathy.
Lu Z, He S, Jiang J, Zhuang L, Wang Y, Yang G, Jiang X, Nie Y, Fu J, Zhang X, Lu Y, Bian X, Chang HC, Xiong Z, Huang X, Liu Z, Sun Q. Lu Z, et al. Mol Ther. 2022 Jun 1;30(6):2163-2175. doi: 10.1016/j.ymthe.2022.03.001. Epub 2022 Mar 11. Mol Ther. 2022. PMID: 35283272 Free PMC article. - Roles of Mso1 and the SM protein Sec1 in efficient vesicle fusion during fission yeast cytokinesis.
Gerien KS, Zhang S, Russell AC, Zhu YH, Purde V, Wu JQ. Gerien KS, et al. Mol Biol Cell. 2020 Jul 15;31(15):1570-1583. doi: 10.1091/mbc.E20-01-0067. Epub 2020 May 20. Mol Biol Cell. 2020. PMID: 32432970 Free PMC article. - The Sec1/Munc18 (SM) protein Vps45 is involved in iron uptake, mitochondrial function and virulence in the pathogenic fungus Cryptococcus neoformans.
Caza M, Hu G, Nielson ED, Cho M, Jung WH, Kronstad JW. Caza M, et al. PLoS Pathog. 2018 Aug 2;14(8):e1007220. doi: 10.1371/journal.ppat.1007220. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30071112 Free PMC article. - SM protein Munc18-2 facilitates transition of Syntaxin 11-mediated lipid mixing to complete fusion for T-lymphocyte cytotoxicity.
Spessott WA, Sanmillan ML, McCormick ME, Kulkarni VV, Giraudo CG. Spessott WA, et al. Proc Natl Acad Sci U S A. 2017 Mar 14;114(11):E2176-E2185. doi: 10.1073/pnas.1617981114. Epub 2017 Mar 6. Proc Natl Acad Sci U S A. 2017. PMID: 28265073 Free PMC article.
References
- Brennwald, P., B. Kearns, K. Champion, S. Keranen, V. Bankaitis, and P. Novick. 1994. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 79:245–258. - PubMed
- Burke, D., D. Dawson, and T. Stearns, and Cold Spring Harbor Laboratory. 2000. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. NY. 205 pp.
- Cowles, C.R., S.D. Emr, and B.F. Horazdovsky. 1994. Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J. Cell Sci. 107:3449–3459. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases