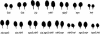SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis - PubMed (original) (raw)
SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis
Angela Peragine et al. Genes Dev. 2004.
Abstract
Higher plants undergo a transition from a juvenile to an adult phase of vegetative development prior to flowering. Screens for mutants that undergo this transition precociously produced alleles of two genes required for posttranscriptional gene silencing (PTGS)--SUPPRESSOR OF GENE SILENCING3 (SGS3) and SUPPRESSOR OF GENE SILENCING2(SGS2)/SILENCING DEFECTIVE1 (SDE1)/RNA-DEPENDENT POLYMERASE6 (RDR6). Loss-of-function mutations in these genes have a phenotype similar to that of mutations in the Argonaute gene ZIPPY (ZIP). Epistasis analysis suggests that ZIP, SGS3, SGS2/SDE1/RDR6, and the putative miRNA export receptor, HASTY (HST), operate in the same pathway(s). Microarray analysis revealed a small number of genes whose mRNA is increased in ZIP, SGS3, and SGS2/SDE1/RDR6 mutants, as well as genes that are up-regulated in SGS3 and SGS2/SDE1/RDR6 mutants, but not in ZIP mutants. One of these latter genes (At5g18040) is silenced posttranscriptionally in trans by the sRNA255 family of endogenous, noncoding, small interfering RNAs (siRNAs). The increase in At5g18040 mRNA in SGS3 and SGS2/SDE1/RDR6 mutants is attributable to the absence of sRNA255-like siRNAs in these mutants. These results demonstrate a role for endogenous siRNAs in the regulation of gene expression, and suggest that PTGS plays a central role in the temporal control of shoot development in plants.
Copyright 2004 Cold Spring Harbor Laboratory Press
Figures
Figure 1.
The morphology of wild-type Columbia (Col), zip-1, sgs3-11, and rdr6-11. (A) Thirteen-day-old plants. The first few leaves of mutant plants are elongated and curl downward. (B) Flowers. Mutant plants have variable seed sets because stamens frequently fail to contact the stigma. (C) Scanning electron micrographs of carpels with one valve and seeds removed. Mutant carpels have a split septum and produce stigmatic tissue in the middle of the septum at the apical end of the carpels.
Figure 2.
The morphology of the first two rosette leaves of wild-type (Col) and mutant plants.
Figure 3.
The effect of zip-1, sgs3-11, and rdr6-11 on posttranscriptional silencing of the L1 (35S::GUS) transgene. zip-1 does not interfere with the silencing of L1, but sgs3-11 and rdr6-11 prevent silencing in all fully expanded rosette leaves.
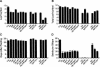
Figure 4.
The phenotype of wild-type (Col) and mutant plants. (A) First leaf with abaxial trichomes. (B) Total number of leaves in the rosette. (C) Time to production of the first open flower. (D) Number of seeds per silique; 25-35 plants were measured for each genotype. Error bars indicate standard error of the mean.
Figure 5.
The phenotype of zip-1, sgs3-11, rdr6-11, hst-1, and sqn-1, and plants homozygous for combinations of these mutations (13-day-old plants).
Figure 6.
The effect of zip-1, sgs3-11, and rdr6-11 on the accumulation of mRNAs, miRNAs, and siRNAs. (A) mRNA of ARF3/ETTIN, ARF4, and SPL3 accumulates in zip, sgs3-11, and rdr6-11. Total RNA from 2-week-old rosettes of plants grown in short days was hybridized with probes to the indicated genes. Actin was used as a loading control. (B) SPL3 mRNA is elevated in zip-1 at 15 and 18 d after planting. Blot of total RNA from shoot apices of wild-type and zip-1 rosettes hybridized with a probe to SPL3. 18S ribosomal RNA was used as a loading control. (C) The accumulation of miRNAs in zip-1, sgs3-11, and rdr6-11. Blots of low-molecular-weight RNA from 2-week-old mutant and wild-type plants hybridized with probes complementary to the functional strand of the indicated miRNAs. (D) The effect of zip-1, sgs3-11, and rdr6-11 on the level of endogenous sRNAs. Blots of low-molecular-weight RNA from 2-week-old mutant and wild-type plants hybridized with probes to the indicated sRNAs. U6 was used as a loading control. The intensity of each signal was measured using NIH Image; the ratio of this signal relative to wild-type is indicated below each image.
Figure 7.
SGS3 and RDR6 repress gene expression by promoting the production of _trans_-acting siRNAs. (A) Genes affected by sgs3-11 and rdr6-11, but not by zip-1. Total RNA from 2-week-old rosettes of plants grown in short days hybridized with probes to the indicated genes. Actin was used as a loading control. The intensity of each signal was measured using NIH Image; the ratio of this signal relative to wild-type is indicated below each image. (B) The genomic structure of At5g18040 and the location and sequence of the cognate site of sRNA255, sRNA752 and sRNA289, and sRNA850. UTRs are black, the coding region is blue, and the sRNA255 cognate sequence is red. Sequencing of nine 5′ RACE clones revealed that At5g18040 is cleaved in the middle of this cognate site. (C) 5′RACE products from Col, sgs3-11, and rdr6-11 2-week-old rosettes. This reaction gives one major product in Col (arrow-head) that is missing in sgs3-11 and rdr6-11. 5′RACE of SCL6-IV, which is cleaved by miR171, was used as a control. (D) sgs3-11, rdr6-11, and dcl1-7 block the production of sRNA255-like RNAs. Blot of low-molecular-weight RNA from 2-week-old plants hybridized with probes complementary to the indicated sRNAs. (E) _sRNA255_-like RNAs are found in three clusters on two different chromosomes. Arrows indicate the 5′-to-3′ orientation of the sRNA sequence. Red arrows are in direct orientation and blue arrows are in reverse orientation to the genomic DNA sequence.
Similar articles
- Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants.
Muangsan N, Beclin C, Vaucheret H, Robertson D. Muangsan N, et al. Plant J. 2004 Jun;38(6):1004-14. doi: 10.1111/j.1365-313X.2004.02103.x. Plant J. 2004. PMID: 15165191 - Genetic interaction between the AS1-AS2 and RDR6-SGS3-AGO7 pathways for leaf morphogenesis.
Xu L, Yang L, Pi L, Liu Q, Ling Q, Wang H, Poethig RS, Huang H. Xu L, et al. Plant Cell Physiol. 2006 Jul;47(7):853-63. doi: 10.1093/pcp/pcj057. Epub 2006 May 13. Plant Cell Physiol. 2006. PMID: 16699177 - DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7.
Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H. Adenot X, et al. Curr Biol. 2006 May 9;16(9):927-32. doi: 10.1016/j.cub.2006.03.035. Curr Biol. 2006. PMID: 16682354 - MicroRNA-dependent trans-acting siRNA production.
Vaucheret H. Vaucheret H. Sci STKE. 2005 Sep 6;2005(300):pe43. doi: 10.1126/stke.3002005pe43. Sci STKE. 2005. PMID: 16145017 Review. - The function of RNAi in plant development.
Poethig RS, Peragine A, Yoshikawa M, Hunter C, Willmann M, Wu G. Poethig RS, et al. Cold Spring Harb Symp Quant Biol. 2006;71:165-70. doi: 10.1101/sqb.2006.71.030. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381293 Review.
Cited by
- Cascading cis-cleavage on transcript from trans-acting siRNA-producing locus 3.
Zhang C, Li G, Wang J, Zhu S, Li H. Zhang C, et al. Int J Mol Sci. 2013 Jul 12;14(7):14689-99. doi: 10.3390/ijms140714689. Int J Mol Sci. 2013. PMID: 23857062 Free PMC article. - Enhanced microRNA accumulation through stemloop-adjacent introns.
Schwab R, Speth C, Laubinger S, Voinnet O. Schwab R, et al. EMBO Rep. 2013 Jul;14(7):615-21. doi: 10.1038/embor.2013.58. Epub 2013 May 10. EMBO Rep. 2013. PMID: 23661080 Free PMC article. - Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants.
Moreno AB, Martínez de Alba AE, Bardou F, Crespi MD, Vaucheret H, Maizel A, Mallory AC. Moreno AB, et al. Nucleic Acids Res. 2013 Apr;41(8):4699-708. doi: 10.1093/nar/gkt152. Epub 2013 Mar 12. Nucleic Acids Res. 2013. PMID: 23482394 Free PMC article. - The Universally Conserved Residues Are Not Universally Required for Stable Protein Expression or Functions of Cryptochromes.
Liu H, Su T, He W, Wang Q, Lin C. Liu H, et al. Mol Biol Evol. 2020 Feb 1;37(2):327-340. doi: 10.1093/molbev/msz217. Mol Biol Evol. 2020. PMID: 31550045 Free PMC article. - RNAi-Based Antiviral Innate Immunity in Plants.
Jin L, Chen M, Xiang M, Guo Z. Jin L, et al. Viruses. 2022 Feb 20;14(2):432. doi: 10.3390/v14020432. Viruses. 2022. PMID: 35216025 Free PMC article. Review.
References
- Alonso J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657. - PubMed
- Ambros V., Lee, R.C., Lavanway, A., Williams, P.T., and Jewell, D. 2003b. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13: 807-818. - PubMed
- Banks J.A., Masson, P., and Fedoroff, N. 1988. Molecular mechanisms in the developmental regulation of the maize Suppressor-mutator transposable element. Genes & Dev. 2: 1364-1380. - PubMed
- Beclin C., Boutet, S., Waterhouse, P., and Vaucheret, H. 2002. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12: 684-688. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials