New method for estimating bacterial cell abundances in natural samples by use of sublimation - PubMed (original) (raw)
Comparative Study
New method for estimating bacterial cell abundances in natural samples by use of sublimation
Daniel P Glavin et al. Appl Environ Microbiol. 2004 Oct.
Abstract
We have developed a new method based on the sublimation of adenine from Escherichia coli to estimate bacterial cell counts in natural samples. To demonstrate this technique, several types of natural samples, including beach sand, seawater, deep-sea sediment, and two soil samples from the Atacama Desert, were heated to a temperature of 500 degrees C for several seconds under reduced pressure. The sublimate was collected on a cold finger, and the amount of adenine released from the samples was then determined by high-performance liquid chromatography with UV absorbance detection. Based on the total amount of adenine recovered from DNA and RNA in these samples, we estimated bacterial cell counts ranging from approximately 10(5) to 10(9) E. coli cell equivalents per gram. For most of these samples, the sublimation-based cell counts were in agreement with total bacterial counts obtained by traditional DAPI (4,6-diamidino-2-phenylindole) staining.
Figures
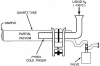
FIG. 1.
The SA used in the heating experiments. The custom-made SA built at the University of California at San Diego was designed specifically for the Lindberg Blue tube furnace and consists of a quartz tube (2.5 by 31 cm) and a Pyrex glass cold finger (1.6 by 15.5 cm), which are sealed together under a vacuum with a clamp and an O-ring. The total cost of the SA was approximately $400. Several vacuum sublimator designs are also available commercially and can be purchased online from Safety Emporium.
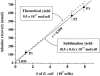
FIG. 2.
The calibration data for the sublimation recovery of adenine from E. coli (solid line). LB medium (LBM) that did not contain E. coli and three separate E. coli pellets (P1, P2, and P3) were heated to 500°C for 30 s in the sublimation apparatus. The total number of E. coli cells was calculated from the mass of each cell pellet given an average wet weight for a single E. coli cell of 9.5 × 10−13 g (18). An average sublimation yield of (8.5 ± 0.4) × 10−17 mol of adenine per cell from E. coli was calculated from the slope of the best-fit line. The uncertainties in the adenine recoveries were based on the standard deviation of the average value of three separate measurements. An error in the sublimation yield of ∼5% was determined from the uncertainty in the slope of the best-fit line. The theoretical yield is also shown for comparison (dashed line).
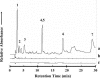
FIG. 3.
The 0- to 30-min region of the reverse-phase HPLC chromatograms of UV absorbance spectra (λ = 260 nm) from sublimed extracts of the (a) Nile Delta deep-sea sediment sample, (b) Atacama Desert subsurface particles, and (c) serpentine blank made after heating the samples in the SA at ∼500°C for 30 s. UV absorbance measurements for the sublimed sand, seawater, and Atacama Desert fine surface particles were also carried out but are not shown here. Peak identifications: (1) HCl front, (2) cytosine, (3) uracil, (4, 5) guanine/hypoxanthine, (6) thymine, and (7) adenine.
References
- Ammerman, J. W., and F. Azam. 1981. Dissolved cAMP in the sea and uptake of cAMP by marine bacteria. Mar. Ecol. Prog. Ser. 5:85-89.
- Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heidal. 1989. High abundance of viruses found in aquatic environments. Nature (London) 340:467-468. - PubMed
- Bernard, L., H. Schäfer, F. Joux, C. Courties, G. Muyzer, and P. Lebaron. 2000. Genetic diversity of total, active and culturable marine bacteria in coastal seawater. Aquatic Microb. Ecol. 23:1-11.
- Cann, A. J. 2001. Principles of molecular virology. Academic Press, San Diego, Calif.
- Cole, S., and I. Saint-Girons. 1999. Bacterial genomes—all shapes and sizes, p. 35-62. In R. Charlebois (ed.), Organization of the prokaryotic genome. ASM Press, Washington, D.C.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources