Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia - PubMed (original) (raw)
Comparative Study
doi: 10.1086/425871. Epub 2004 Oct 1.
Affiliations
- PMID: 15467980
- PMCID: PMC1182158
- DOI: 10.1086/425871
Comparative Study
Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia
Malliya Gounder Palanichamy et al. Am J Hum Genet. 2004 Dec.
Abstract
To resolve the phylogeny of the autochthonous mitochondrial DNA (mtDNA) haplogroups of India and determine the relationship between the Indian and western Eurasian mtDNA pools more precisely, a diverse subset of 75 macrohaplogroup N lineages was chosen for complete sequencing from a collection of >800 control-region sequences sampled across India. We identified five new autochthonous haplogroups (R7, R8, R30, R31, and N5) and fully characterized the autochthonous haplogroups (R5, R6, N1d, U2a, U2b, and U2c) that were previously described only by first hypervariable segment (HVS-I) sequencing and coding-region restriction-fragment-length polymorphism analysis. Our findings demonstrate that the Indian mtDNA pool, even when restricted to macrohaplogroup N, harbors at least as many deepest-branching lineages as the western Eurasian mtDNA pool. Moreover, the distribution of the earliest branches within haplogroups M, N, and R across Eurasia and Oceania provides additional evidence for a three-founder-mtDNA scenario and a single migration route out of Africa.
Figures
Figure 1
Phylogenetic tree of 75 Indian complete mtDNA sequences. Parts A, B, C, and D of the figure are the phylogenies of the N, pre-HV and JT, U, and Indian autochthonous R haplogroups, respectively. Mutations are scored relative to the rCRS (Andrews et al. 1999). Indian populations: A = Bhargava, B = Chaturvedi, C = Brahmin, R = Reddy, S = Khasi, SW = Rajbhansi, T = Thogataveera. Twenty-five additional complete sequences were taken from the literature (Finnilä et al. ; Maca-Meyer et al. ; Taylor et al. ; Derbeneva et al. ; Herrnstadt et al. , ; Ingman and Gyllensten ; Mishmar et al. ; Coble et al. 2004), and we referred to particular samples from these articles by SF, NM, RT, OD, CH, IG, DM, and MC, respectively, followed by “#” and the original sample code. Suffixes A, C, G, and T indicate transversions; “d” denotes a deletion, and a plus sign (+) denotes an insertion; recurrent mutations are underlined; “h” indicates heteroplasmy; and italics highlight likely oversights. Mutations in the single reported haplogroup N1a lineage labeled by an asterisk (*) are our reconstruction. The linkage between coding- and control-region mutations in the new haplogroup U2d is tentative. Since the variation at 16519 is extremely hypervariable, only the most parsimonious solution is offered here. For haplogroups H and U5, see articles by Loogväli et al. (2004) and Tambets et al. (2004), respectively.
Figure 1
Phylogenetic tree of 75 Indian complete mtDNA sequences. Parts A, B, C, and D of the figure are the phylogenies of the N, pre-HV and JT, U, and Indian autochthonous R haplogroups, respectively. Mutations are scored relative to the rCRS (Andrews et al. 1999). Indian populations: A = Bhargava, B = Chaturvedi, C = Brahmin, R = Reddy, S = Khasi, SW = Rajbhansi, T = Thogataveera. Twenty-five additional complete sequences were taken from the literature (Finnilä et al. ; Maca-Meyer et al. ; Taylor et al. ; Derbeneva et al. ; Herrnstadt et al. , ; Ingman and Gyllensten ; Mishmar et al. ; Coble et al. 2004), and we referred to particular samples from these articles by SF, NM, RT, OD, CH, IG, DM, and MC, respectively, followed by “#” and the original sample code. Suffixes A, C, G, and T indicate transversions; “d” denotes a deletion, and a plus sign (+) denotes an insertion; recurrent mutations are underlined; “h” indicates heteroplasmy; and italics highlight likely oversights. Mutations in the single reported haplogroup N1a lineage labeled by an asterisk (*) are our reconstruction. The linkage between coding- and control-region mutations in the new haplogroup U2d is tentative. Since the variation at 16519 is extremely hypervariable, only the most parsimonious solution is offered here. For haplogroups H and U5, see articles by Loogväli et al. (2004) and Tambets et al. (2004), respectively.
Figure 1
Phylogenetic tree of 75 Indian complete mtDNA sequences. Parts A, B, C, and D of the figure are the phylogenies of the N, pre-HV and JT, U, and Indian autochthonous R haplogroups, respectively. Mutations are scored relative to the rCRS (Andrews et al. 1999). Indian populations: A = Bhargava, B = Chaturvedi, C = Brahmin, R = Reddy, S = Khasi, SW = Rajbhansi, T = Thogataveera. Twenty-five additional complete sequences were taken from the literature (Finnilä et al. ; Maca-Meyer et al. ; Taylor et al. ; Derbeneva et al. ; Herrnstadt et al. , ; Ingman and Gyllensten ; Mishmar et al. ; Coble et al. 2004), and we referred to particular samples from these articles by SF, NM, RT, OD, CH, IG, DM, and MC, respectively, followed by “#” and the original sample code. Suffixes A, C, G, and T indicate transversions; “d” denotes a deletion, and a plus sign (+) denotes an insertion; recurrent mutations are underlined; “h” indicates heteroplasmy; and italics highlight likely oversights. Mutations in the single reported haplogroup N1a lineage labeled by an asterisk (*) are our reconstruction. The linkage between coding- and control-region mutations in the new haplogroup U2d is tentative. Since the variation at 16519 is extremely hypervariable, only the most parsimonious solution is offered here. For haplogroups H and U5, see articles by Loogväli et al. (2004) and Tambets et al. (2004), respectively.
Figure 1
Phylogenetic tree of 75 Indian complete mtDNA sequences. Parts A, B, C, and D of the figure are the phylogenies of the N, pre-HV and JT, U, and Indian autochthonous R haplogroups, respectively. Mutations are scored relative to the rCRS (Andrews et al. 1999). Indian populations: A = Bhargava, B = Chaturvedi, C = Brahmin, R = Reddy, S = Khasi, SW = Rajbhansi, T = Thogataveera. Twenty-five additional complete sequences were taken from the literature (Finnilä et al. ; Maca-Meyer et al. ; Taylor et al. ; Derbeneva et al. ; Herrnstadt et al. , ; Ingman and Gyllensten ; Mishmar et al. ; Coble et al. 2004), and we referred to particular samples from these articles by SF, NM, RT, OD, CH, IG, DM, and MC, respectively, followed by “#” and the original sample code. Suffixes A, C, G, and T indicate transversions; “d” denotes a deletion, and a plus sign (+) denotes an insertion; recurrent mutations are underlined; “h” indicates heteroplasmy; and italics highlight likely oversights. Mutations in the single reported haplogroup N1a lineage labeled by an asterisk (*) are our reconstruction. The linkage between coding- and control-region mutations in the new haplogroup U2d is tentative. Since the variation at 16519 is extremely hypervariable, only the most parsimonious solution is offered here. For haplogroups H and U5, see articles by Loogväli et al. (2004) and Tambets et al. (2004), respectively.
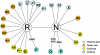
Figure 2
Subcontinental ancestry of the most basal Eurasian/Oceanian branches of the mtDNA phylogeny. South Asian and western Eurasian haplogroups are defined as described in the present study; for East Asian haplogroups, see Kong et al. (2003); for haplogroup P, see Forster et al. (2001); O and S are newly defined here on the basis of the data from Ingman and Gyllensten (2003). Potential coalescences based only on a single highly variable site are disregarded.
Similar articles
- Carriers of Mitochondrial DNA Macrohaplogroup N Lineages Reached Australia around 50,000 Years Ago following a Northern Asian Route.
Fregel R, Cabrera V, Larruga JM, Abu-Amero KK, González AM. Fregel R, et al. PLoS One. 2015 Jun 8;10(6):e0129839. doi: 10.1371/journal.pone.0129839. eCollection 2015. PLoS One. 2015. PMID: 26053380 Free PMC article. - The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes.
Sun C, Kong QP, Palanichamy MG, Agrawal S, Bandelt HJ, Yao YG, Khan F, Zhu CL, Chaudhuri TK, Zhang YP. Sun C, et al. Mol Biol Evol. 2006 Mar;23(3):683-90. doi: 10.1093/molbev/msj078. Epub 2005 Dec 16. Mol Biol Evol. 2006. PMID: 16361303 - Carriers of human mitochondrial DNA macrohaplogroup M colonized India from southeastern Asia.
Marrero P, Abu-Amero KK, Larruga JM, Cabrera VM. Marrero P, et al. BMC Evol Biol. 2016 Nov 10;16(1):246. doi: 10.1186/s12862-016-0816-8. BMC Evol Biol. 2016. PMID: 27832758 Free PMC article. - A comprehensive review of HVS-I mitochondrial DNA variation of 19 Iranian populations.
Amjadi M, Hayatmehr Z, Egyed B, Tavallaei M, Szécsényi-Nagy A. Amjadi M, et al. Ann Hum Genet. 2024 May;88(3):259-277. doi: 10.1111/ahg.12544. Epub 2023 Dec 31. Ann Hum Genet. 2024. PMID: 38161274 Review. - Complex genetic origin of Indian populations and its implications.
Tamang R, Singh L, Thangaraj K. Tamang R, et al. J Biosci. 2012 Nov;37(5):911-9. doi: 10.1007/s12038-012-9256-9. J Biosci. 2012. PMID: 23107926 Review.
Cited by
- A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios.
Sahoo S, Singh A, Himabindu G, Banerjee J, Sitalaximi T, Gaikwad S, Trivedi R, Endicott P, Kivisild T, Metspalu M, Villems R, Kashyap VK. Sahoo S, et al. Proc Natl Acad Sci U S A. 2006 Jan 24;103(4):843-8. doi: 10.1073/pnas.0507714103. Epub 2006 Jan 13. Proc Natl Acad Sci U S A. 2006. PMID: 16415161 Free PMC article. - Austro-Asiatic tribes of Northeast India provide hitherto missing genetic link between South and Southeast Asia.
Reddy BM, Langstieh BT, Kumar V, Nagaraja T, Reddy AN, Meka A, Reddy AG, Thangaraj K, Singh L. Reddy BM, et al. PLoS One. 2007 Nov 7;2(11):e1141. doi: 10.1371/journal.pone.0001141. PLoS One. 2007. PMID: 17989774 Free PMC article. - The peopling of Europe from the mitochondrial haplogroup U5 perspective.
Malyarchuk B, Derenko M, Grzybowski T, Perkova M, Rogalla U, Vanecek T, Tsybovsky I. Malyarchuk B, et al. PLoS One. 2010 Apr 21;5(4):e10285. doi: 10.1371/journal.pone.0010285. PLoS One. 2010. PMID: 20422015 Free PMC article. - Mitochondrial DNA structure in the Arabian Peninsula.
Abu-Amero KK, Larruga JM, Cabrera VM, González AM. Abu-Amero KK, et al. BMC Evol Biol. 2008 Feb 12;8:45. doi: 10.1186/1471-2148-8-45. BMC Evol Biol. 2008. PMID: 18269758 Free PMC article. - Phylogeographic distribution of mitochondrial DNA macrohaplogroup M in India.
Maji S, Krithika S, Vasulu TS. Maji S, et al. J Genet. 2009 Apr;88(1):127-39. doi: 10.1007/s12041-009-0020-3. J Genet. 2009. PMID: 19417557
References
Electronic-Database Information
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for accession numbers AY713976–AY714050)
References
- Allard MW, Miller K, Wilson MR, Monson KL, Budowle B (2002) Characterization of the Caucasian haplogroups present in the SWGDAM forensic mtDNA data set for 1771 human control region sequences. J Forensic Sci 47:1215–1223 - PubMed
- Allard MW, Wilson MR, Monson KL, Budowle B (2004) Control region sequences for East Asian individuals in the Scientific Working Group on DNA Analysis Methods forensic mtDNA data set. Legal Med 6:11–24 - PubMed
- Al-Zahery N, Semino O, Benuzzi G, Magri C, Passarino G, Torroni A, Santachiara-Benerecetti AS (2003) Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol 28:458–472 - PubMed
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 - PubMed
- Baig M, Khan A, Kulkarni K. Mitochondrial DNA diversity in tribal and caste groups of Maharashtra (India) and its implication on their genetic origins. Ann Hum Genet (in press) - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases