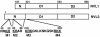NVL2 is a nucleolar AAA-ATPase that interacts with ribosomal protein L5 through its nucleolar localization sequence - PubMed (original) (raw)
NVL2 is a nucleolar AAA-ATPase that interacts with ribosomal protein L5 through its nucleolar localization sequence
Masami Nagahama et al. Mol Biol Cell. 2004 Dec.
Abstract
NVL (nuclear VCP-like protein), a member of the AAA-ATPase family, is known to exist in two forms with N-terminal extensions of different lengths in mammalian cells. Here, we show that they are localized differently in the nucleus; NVL2, the major species, is mainly present in the nucleolus, whereas NVL1 is nucleoplasmic. Mutational analysis demonstrated the presence of two nuclear localization signals in NVL2, one of which is shared with NVL1. In addition, a nucleolar localization signal was found to exist in the N-terminal extra region of NVL2. The nucleolar localization signal is critical for interaction with ribosomal protein L5, which was identified as a specific interaction partner of NVL2 on yeast two-hybrid screening. The interaction of NVL2 with L5 is ATP-dependent and likely contributes to the nucleolar translocation of NVL2. The physiological implication of this interaction was suggested by the finding that a dominant negative NVL2 mutant inhibits ribosome biosynthesis, which is known to take place in the nucleolus.
Figures
Figure 1.
Schematic representation of human NVL1 and NVL2. The numbering of amino acids starts from the initiation methionine of NVL2. The translation of NVL1 starts from the residue corresponding to the second methionine (residue 107) of NVL2. The amino acid sequences of potential nuclear and nucleolar localization signals are indicated. The residues mutated in this study are underlined, and named M1, M2, M3, and M4, respectively.
Figure 2.
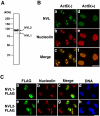
Differential subnuclear localization of NVL isoforms. (A) A 293T cell lysate was analyzed by Western blotting using an anti-NVL antibody. The molecular size markers are indicated on the left. (B) HeLa cells were untreated or treated with ActD (50 ng/ml) for 3 h. The localization of endogenous NVL proteins and nucleolin was determined by fluorescence microscopy after double staining with anti-NVL and anti-nucleolin antibodies. (C) Immunofluorescence analysis of the localization of NVL isoforms. HeLa cells were transiently transfected with FLAG-tagged NVL1 and NVL2. After 20 h, the cells were fixed and double-stained with anti-FLAG and anti-nucleolin antibodies. Nuclei were visualized with DAPI (DNA). Bars, 20 μm.
Figure 3.
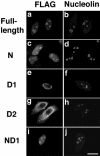
Subcellular localization of each domain of NVL2. HeLa cells were transiently transfected with full-length FLAG-tagged NVL2 or a truncation mutant representing the N- (residues 1–220), D1- (residues 221–553), D2- (residues 554–856), or ND1- (N- and D1-) domain. After 20 h, the cells were fixed and double-stained with anti-FLAG and anti-nucleolin antibodies. Bars, 20 μm.
Figure 4.
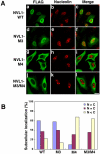
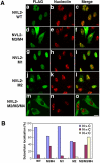
Identification of a nuclear localization signal in NVL1. (A) HeLa cells were transiently transfected with the FLAG-tagged wild-type (WT) NVL1, NVL1-M3, NVL1-M4, and NVL1-M3/M4. After 20 h, the cells were fixed and double-stained with anti-FLAG and anti-nucleolin antibodies. Bars, 20 μm. (B) Quantitative analysis of the results in A. The cellular localization of the indicated constructs was scored as follows: cells showing more intense nuclear than cytoplasmic staining (N > C), cells showing a nearly equal distribution between the cytoplasm and nucleus (N = C), and cells showing predominantly cytoplasmic staining (N < C). At least 100 cells were scored for each construct.
Figure 5.
Identification of nuclear and nucleolar localization signals in NVL2. (A) HeLa cells were transiently transfected with the FLAG-tagged wild-type (WT) NVL2, NVL2-M3/M4, NVL2-M1, NVL2-M2, and NVL2-M2/M3/M4. After 20 h, the cells were fixed and double-stained with anti-FLAG and anti-nucleolin antibodies. Bars, 20 μm. (B) Quantitative analysis of the results in A. At least 100 cells were scored for each construct.
Figure 6.
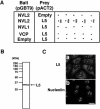
L5 is an interaction partner of NVL2. (A) For yeast two-hybrid analysis, S. cerevisiae Y190 cells were transformed with bait vector pGBT9 containing NVL2, NVL1, or VCP/p97, or pGBT9 alone as a control (Empty), and prey vector pACT2 containing L5 (residues 81–297) or pACT2 alone as a control (Empty). The interaction was examined by monitoring β-galactosidase activity on a filter. Five independent transformants were examined for each pair of constructs. (B) A 293T cell lysate was analyzed by Western blotting using an anti-L5 antibody. The molecular size markers are indicated on the left. (C) The localization of endogenous L5 and nucleolin was determined by fluorescence microscopy after double-staining with anti-L5 and anti-nucleolin antibodies. Bars, 20 μm.
Figure 7.
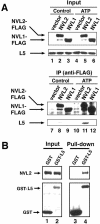
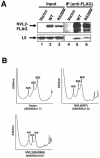
NVL2 interacts with L5 in an ATP-dependent manner. (A) 293T cells were transfected with FLAG-tagged forms of NVL1 or NVL2. After 24 h, cell lysates were subjected to immunoprecipitation with an anti-FLAG antibody in the presence or absence (Control) of ATP. The immunoprecipitates (IP) and 2% of the starting material (Input) were resolved by SDS-PAGE and then analyzed by Western blotting. (B) A cell lysate of 293T cells expressing GST-L5 or GST as a control was subjected to GST pull-down assay with glutathione beads. ATP was included during the incubation. The proteins bound to glutathione beads (Pull-down) and 2% of the starting material (Input) were resolved by SDS-PAGE and then analyzed by Western blotting.
Figure 8.
The NoLS of NVL2 is required for its interaction with L5. (A) For yeast two-hybrid analysis, S. cerevisiae Y190 cells were transformed with bait vector pGBT9 containing the indicated NVL2 domain, or pGBT9 alone as a control (Empty), and prey vector pACT2 containing L5 (residues 81–297) or pACT2 alone as a control (Empty). The interaction was examined by monitoring β-galactosidase activity on a filter. Five independent transformants were examined for each pair of constructs. full, full-length. (B) 293T cells were transfected with the indicated FLAG-tagged mutant of NVL1 or NVL2. After 24 h, cell lysates were subjected to immunoprecipitation with an anti-FLAG antibody. ATP was included during the incubation. The immunoprecipitates (IP) and 2% of the starting material (Input) were resolved by SDS-PAGE and then analyzed by Western blotting.
Figure 9.
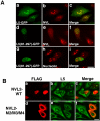
N-terminally truncated L5(81–297) relocalizes NVL2 from the nucleolus to nucleoplasmic punctate structures. (A) HeLa cells were transiently transfected with L5-GFP or L5(81–297)-GFP. After 20 h, the cells were fixed and stained with an anti-NVL antibody (panels b and e) or anti-nucleolin antibody (panel h). L5-GFP (panel a) or L5(81–297)-GFP (panels d and g) was visualized directly. Merged images are shown in the right panels. (B) HeLa cells were transfected with FLAG-tagged forms of the wild-type (WT) NVL2 and the NVL2-M2/M3/M4 mutant. After 20 h, the cells were fixed and double-stained with an anti-FLAG antibody and anti-L5 antibody. Merged images are shown in the right panels. Bars, 20 μm.
Figure 10.
Overexpression of the K628M mutant of NVL2 impairs 60S subunit biogenesis. (A) 293T cells were transfected with FLAG-tagged forms of the wild-type NVL2 (WT) and the K628M mutant. After 24 h, cell lysates were subjected to immunoprecipitation with an anti-FLAG antibody in the presence of ATP. The immunoprecipitates (IP) and 2% of the starting material (Input) were resolved by SDS-PAGE and then analyzed by Western blotting. (B) The ribosome profiles (A254 nm) on sucrose gradient centrifugation of extracts of cells transfected with the control vector and the expression plasmids for the FLAG-tagged wild-type NVL2 (WT) and K628M mutant are shown. The positions of the 40S, 60S, and 80S ribosomal subunits are indicated.
Similar articles
- The AAA-ATPase NVL2 is a component of pre-ribosomal particles that interacts with the DExD/H-box RNA helicase DOB1.
Nagahama M, Yamazoe T, Hara Y, Tani K, Tsuji A, Tagaya M. Nagahama M, et al. Biochem Biophys Res Commun. 2006 Aug 4;346(3):1075-82. doi: 10.1016/j.bbrc.2006.06.017. Epub 2006 Jun 12. Biochem Biophys Res Commun. 2006. PMID: 16782053 - AAA-ATPase NVL2 acts on MTR4-exosome complex to dissociate the nucleolar protein WDR74.
Hiraishi N, Ishida Y, Nagahama M. Hiraishi N, et al. Biochem Biophys Res Commun. 2015 Nov 20;467(3):534-40. doi: 10.1016/j.bbrc.2015.09.160. Epub 2015 Oct 8. Biochem Biophys Res Commun. 2015. PMID: 26456651 - Structure and function of the N-terminal nucleolin binding domain of nuclear valosin-containing protein-like 2 (NVL2) harboring a nucleolar localization signal.
Fujiwara Y, Fujiwara K, Goda N, Iwaya N, Tenno T, Shirakawa M, Hiroaki H. Fujiwara Y, et al. J Biol Chem. 2011 Jun 17;286(24):21732-41. doi: 10.1074/jbc.M110.174680. Epub 2011 Apr 7. J Biol Chem. 2011. PMID: 21474449 Free PMC article. - [Regulation of ribosome assembly by molecular chaperones --functional analysis of nucleolar AAA-ATPase NVL2].
Nagahama M. Nagahama M. Seikagaku. 2013 Oct;85(10):880-8. Seikagaku. 2013. PMID: 24392587 Review. Japanese. No abstract available. - Ribosomal Proteins Control or Bypass p53 during Nucleolar Stress.
Russo A, Russo G. Russo A, et al. Int J Mol Sci. 2017 Jan 12;18(1):140. doi: 10.3390/ijms18010140. Int J Mol Sci. 2017. PMID: 28085118 Free PMC article. Review.
Cited by
- Telomere lengthening and other functions of telomerase.
Rubtsova MP, Vasilkova DP, Malyavko AN, Naraikina YV, Zvereva MI, Dontsova OA. Rubtsova MP, et al. Acta Naturae. 2012 Apr;4(2):44-61. Acta Naturae. 2012. PMID: 22872811 Free PMC article. - Detecting differential transcript usage in complex diseases with SPIT.
Erdogdu B, Varabyou A, Hicks SC, Salzberg SL, Pertea M. Erdogdu B, et al. bioRxiv [Preprint]. 2023 Jul 10:2023.07.10.548289. doi: 10.1101/2023.07.10.548289. bioRxiv. 2023. PMID: 37503064 Free PMC article. Updated. Preprint. - Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7.
Lo YH, Sobhany M, Hsu AL, Ford BL, Krahn JM, Borgnia MJ, Stanley RE. Lo YH, et al. Nat Commun. 2019 Jan 31;10(1):513. doi: 10.1038/s41467-019-08373-0. Nat Commun. 2019. PMID: 30705282 Free PMC article. - Localization and Functional Roles of Components of the Translation Apparatus in the Eukaryotic Cell Nucleus.
Kachaev ZM, Ivashchenko SD, Kozlov EN, Lebedeva LA, Shidlovskii YV. Kachaev ZM, et al. Cells. 2021 Nov 19;10(11):3239. doi: 10.3390/cells10113239. Cells. 2021. PMID: 34831461 Free PMC article. Review. - Structural Analysis Reveals Features of Ribosome Assembly Factor Nsa1/WDR74 Important for Localization and Interaction with Rix7/NVL2.
Lo YH, Romes EM, Pillon MC, Sobhany M, Stanley RE. Lo YH, et al. Structure. 2017 May 2;25(5):762-772.e4. doi: 10.1016/j.str.2017.03.008. Epub 2017 Apr 13. Structure. 2017. PMID: 28416111 Free PMC article.
References
- Bernardi, R., Scaglioni, P.P., Bergmann, S., Horn, H.F., Vousden, K.H., and Pandolfi, P.P. (2004). PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 6, 665-672. - PubMed
- Carmo-Fonseca, M., Mendes-Soares, L., and Campos, I. (2000). To be or not to be in the nucleolus. Nat. Cell Biol. 2, E107-E112. - PubMed
- Claussen, M., Rudt, F., and Pieler, T. (1999). Functional modules in ribosomal protein L5 for ribonucleoprotein complex formation and nucleocytoplasmic transport. J. Biol. Chem. 274, 33951-33958. - PubMed
- Dingwall, C., and Laskey, R.A. (1991). Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 16, 478-481. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous