The wrickkened pathways of FGF23, MEPE and PHEX - PubMed (original) (raw)
Review
The wrickkened pathways of FGF23, MEPE and PHEX
Peter S N Rowe. Crit Rev Oral Biol Med. 2004.
Abstract
The last 350 years since the publication of the first medical monograph on rickets (old English term wrickken) (Glisson et al., 1651) have seen spectacular advances in our understanding of mineral-homeostasis. Seminal and exciting discoveries have revealed the roles of PTH, vitamin D, and calcitonin in regulating calcium and phosphate, and maintaining healthy teeth and skeleton. However, it is clear that the PTH/Vitamin D axis does not account for the entire picture, and a new bone-renal metabolic milieu has emerged, implicating a novel set of matrix proteins, hormones, and Zn-metallopeptidases. The primary defects in X-linked hypophosphatemic rickets (HYP) and autosomal-dominant hypophosphatemic rickets (ADHR) are now identified as inactivating mutations in a Zn-metalloendopeptidase (PHEX) and activating mutations in fibroblast-growth-factor-23 (FGF23), respectively. In oncogenic hypophosphatemic osteomalacia (OHO), several tumor-expressed proteins (MEPE, FGF23, and FRP-4) have emerged as candidate mediators of the bone-renal pathophysiology. This has stimulated the proposal of a global model that takes into account the remarkable similarities between the inherited diseases (HYP and ADHR) and the tumor-acquired disease OHO. In HYP, loss of PHEX function is proposed to result in an increase in uncleaved full-length FGF23 and/or inappropriate processing of MEPE. In ADHR, a mutation in FGF23 results in resistance to proteolysis by PHEX or other proteases and an increase in half-life of full-length phosphaturic FGF23. In OHO, over-expression of FGF23 and/or MEPE is proposed to result in abnormal renal-phosphate handling and mineralization. Although this model is attractive, many questions remain unanswered, suggesting a more complex picture. The following review will present a global hypothesis that attempts to explain the experimental and clinical observations in HYP, ADHR, and OHO, plus diverse mouse models that include the MEPE null mutant, HYP-PHEX transgenic mouse, and MEPE-PHEX double-null-mutant.
Figures
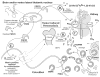
Figure 1
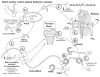
Proposed role for PHEX, MEPE, and the ASARM peptide in mineralization. MEPE and osteoblastic proteases (NEP, ECEL-1/DINE, and cathepsin-D) are markedly up-regulated in murine hyposteoblasts that have defective phex (Jo et al., 2000, 2001; Argiro et al., 2001; Bai et al., 2002; Dubois et al., 2002; Guo et al., 2002; Liu et al., 2003). PHEX/phex protects MEPE from cathepsin-B and general protease degradation and prevents release of the ASARM peptide (Guo et al., 2002). Cathepsin-B is also expressed in the osteoblast (Aisa et al., 1996, 2003). Thus, in HYP/hyp, elevated MEPE (with elevated osteoblastic proteases and loss of PHEX protease protection of MEPE) results in elevated levels of protease-resistant MEPE-ASARM peptide. The MEPE-ASARM peptide inhibits mineralization in vivo, and the osteopontin ASARM peptide potently inhibits calcium oxalate crystallization and crystal growth (Rowe et al., 2000, 2004; Hoyer et al., 2001). Also, the salivary statherin ASARM peptide (see Fig. 2) plays a direct biological role in inhibiting spontaneous precipitation of supersaturated salivary calcium and phosphate and maintaining the mineralization dynamics of tooth enamel (Schlesinger and Hay, 1977; Bennick et al., 1981; Raj et al., 1992; Long et al., 1998). Of related interest, the MEPE knockout, as expected, has accelerated mineralization and increased bone density and bone formation (Gowen et al., 2003). Mineral maturity (mineral crystal size and perfection) throughout all anatomic regions of the osteopontin knock-out mouse bone are also significantly increased (Boskey et al., 2002).
Figure 2
Salivary-statherin & MEPE consensus ASARM motif: mineralization-inhibition and ancestral genes on chromosome 4. MEPE, DMP-1, and the SIBLINGs are related to an ancestral mineralization-gene (salivary statherin) that is also thought to function in the transport of calcium and phosphate in salivary glands. Statherin maps to chromosome 4 in the SIBLING/MEPE region and also contains an ASARM motif. (A) A clustal alignment of the COOH terminal region of human-DMP-1, human-MEPE, mouse-MEPE, and rat-MEPE with human-statherin (62-residue protein). In MEPE and DMP-1, the ASARM peptide occupies the most distal COOH-region of the molecule and is highlighted with a boxed cartouche labeled ‘MEPE ASARM peptide’. The boxed cartouche labeled as ‘statherin-ASARM peptide’ contains the sequence shown to play a biological role in inhibiting spontaneous precipitation of supersaturated salivary calcium and phosphate and maintaining the mineralization dynamics of tooth enamel (Schlesinger and Hay, 1977; Bennick et al., 1981; Raj et al., 1992; Long et al., 1998). Statherin is also thought to function in the transport of calcium and phosphate in salivary glands (Raj et al., 1992). In both MEPE and statherin, cathepsin-B and/or general protease cleavage results in the release of MEPE and statherin protease-resistant ASARM peptides. Both MEPE and statherin ASARM peptides are phosphorylated, protease-resistant, acidic and highly charged molecules with low pI’s. They also share biological properties. For example, a feature of the MEPE ASARM region is the repeat (D) SSES/E sequence. This short sequence has been shown to be a key inhibitor of hydroxyapatite crystal formation and mineralization in salivary statherin (Raj et al., 1992; Long et al., 1998). (B) Schematic presentation of the remarkable clustering of MEPE, DMP-1, statherin, and other SIBLING genes on chromosome 4.
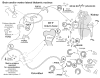
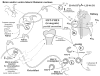
Figure 3
Normal physiology. A scheme representing the proposed dynamic and interactive roles of PHEX, FGF23, MEPE, and matrix proteins in maintaining normal tooth-bone-renal-mineralization and phosphate homeostasis. See text for detailed description (specifically, the sub-paragraph entitled ‘Normal Physiology [Global Hypothesis]’). Also refer to the Table for more details concerning the icons used to represent the diverse molecules, pathways, and tissues.
Figure 4
X-linked rickets (Hyp). A cartoon illustrating the proposed changes in X-linked hypophosphatemic rickets. The primary defect in this disease is the PHEX gene, and this has an impact on renal phosphate handling, vitamin D metabolism, and mineralization. For comparison with normal states, see Fig. 3. See text for detailed description (specifically, the subparagraph entitled ‘X-linked hypophosphatemic rickets [Hyp] [Global Hypothesis]’). Also refer to the Table for more details concerning the icons used to represent the diverse molecules, pathways, and tissues.
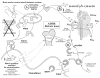
Figure 5
Autosomal-dominant hypophosphatemic rickets (ADHR). Global scheme proposing the changes in autosomal-dominant hypophosphatemic rickets (ADHR). See text for detailed description (specifically, the subparagraph entitled ‘Autosomal-dominant hypophosphatemic rickets [ADHR] [Global Hypothesis]’). Also refer to the Table for more details concerning the icons used to represent the diverse molecules, pathways, and tissues.
Figure 6
Oncogenic hypophosphatemic hypophosphatemia (OHO). Global scheme proposing the changes in oncogenic hypophosphatemic osteomalacia, also known as tumor-induced osteomalacia (OHO). See text for detailed description (specifically, the subparagraph entitled ‘Tumor-induced osteomalacia [OHO] [Global Hypothesis]’). Also refer to the Table for more details concerning the icons used to represent the diverse molecules, pathways, and tissues.
Figure 7
The phex transgenic hyp-mouse: hyp-mice with re-introduced wild-type phex genes have partially corrected mineralization phenotypes and remain hypophosphatemic. See text for a full explanation of the proposed model for the experimental observations (specifically, the subparagraph entitled ‘HYP-PHEX transgenic [partial correction of mineralization phenotype] [Global Hypothesis]’). Also refer to the Table for more details concerning the icons used to represent the diverse molecules, pathways, and tissues.
Similar articles
- Regulation of bone-renal mineral and energy metabolism: the PHEX, FGF23, DMP1, MEPE ASARM pathway.
Rowe PS. Rowe PS. Crit Rev Eukaryot Gene Expr. 2012;22(1):61-86. doi: 10.1615/critreveukargeneexpr.v22.i1.50. Crit Rev Eukaryot Gene Expr. 2012. PMID: 22339660 Free PMC article. Review. - FGF23, PHEX, and MEPE regulation of phosphate homeostasis and skeletal mineralization.
Quarles LD. Quarles LD. Am J Physiol Endocrinol Metab. 2003 Jul;285(1):E1-9. doi: 10.1152/ajpendo.00016.2003. Am J Physiol Endocrinol Metab. 2003. PMID: 12791601 Review. - Distinct roles for intrinsic osteocyte abnormalities and systemic factors in regulation of FGF23 and bone mineralization in Hyp mice.
Liu S, Tang W, Zhou J, Vierthaler L, Quarles LD. Liu S, et al. Am J Physiol Endocrinol Metab. 2007 Dec;293(6):E1636-44. doi: 10.1152/ajpendo.00396.2007. Epub 2007 Sep 11. Am J Physiol Endocrinol Metab. 2007. PMID: 17848631 - Bone proteins PHEX and DMP1 regulate fibroblastic growth factor Fgf23 expression in osteocytes through a common pathway involving FGF receptor (FGFR) signaling.
Martin A, Liu S, David V, Li H, Karydis A, Feng JQ, Quarles LD. Martin A, et al. FASEB J. 2011 Aug;25(8):2551-62. doi: 10.1096/fj.10-177816. Epub 2011 Apr 20. FASEB J. 2011. PMID: 21507898 Free PMC article.
Cited by
- PTH receptor signalling, osteocytes and bone disease induced by diabetes mellitus.
Marino S, Bellido T. Marino S, et al. Nat Rev Endocrinol. 2024 Nov;20(11):661-672. doi: 10.1038/s41574-024-01014-7. Epub 2024 Jul 17. Nat Rev Endocrinol. 2024. PMID: 39020007 Review. - Leveraging FAM features to predict the prognosis of LGG patients and immunotherapy outcome.
Lin L, Yu H, Xie X, Lei Q, Chen X, Su X, Wang X, Zhang S, Yang W. Lin L, et al. Am J Cancer Res. 2024 Jun 15;14(6):2731-2754. doi: 10.62347/GIGO3446. eCollection 2024. Am J Cancer Res. 2024. PMID: 39005680 Free PMC article. - Characteristics of oral health of patients with X-linked hypophosphatemia: case reports and literature review.
Arhar A, Pavlič A, Hočevar L. Arhar A, et al. BDJ Open. 2024 May 31;10(1):42. doi: 10.1038/s41405-024-00223-6. BDJ Open. 2024. PMID: 38821917 Free PMC article. Review. - The Intricacies of Renal Phosphate Reabsorption-An Overview.
Walker V. Walker V. Int J Mol Sci. 2024 Apr 25;25(9):4684. doi: 10.3390/ijms25094684. Int J Mol Sci. 2024. PMID: 38731904 Free PMC article. Review. - Immunolocalization of Enzymes/Membrane Transporters Related to Bone Mineralization in the Metaphyses of the Long Bones of Parathyroid-Hormone-Administered Mice.
Mae T, Hasegawa T, Hongo H, Yamamoto T, Zhao S, Li M, Yamazaki Y, Amizuka N. Mae T, et al. Medicina (Kaunas). 2023 Jun 20;59(6):1179. doi: 10.3390/medicina59061179. Medicina (Kaunas). 2023. PMID: 37374382 Free PMC article.
References
- Aisa MC, Rahman S, Senin U, Maggio D, Russell RG. Cathepsin B activity in normal human osteoblast-like cells and human osteoblastic osteosarcoma cells (MG-63): regulation by interleukin-1 beta and parathyroid hormone. Biochim Biophys Acta. 1996;1290:29–36. - PubMed
- Aisa MC, Beccari T, Costanzi E, Maggio D. Cathepsin B in osteoblasts. Biochim Biophys Acta. 2003;1621:149–159. - PubMed
- Allen HC, Fedarko NS, Jain A, Whybro A, Barker ME, Eastell R, et al. The response of MEPE to long-term and short-term phosphate supplementation in healthy volunteers (abstract) J Bone Miner Res. 2003;18:S116.
- Argiro L, Desbarats M, Glorieux FH, Ecarot B. Mepe, the gene encoding a tumor-secreted protein in oncogenic hypophosphatemic osteomalacia, is expressed in bone. Genomics. 2001;74:342–351. - PubMed
- Aschinberg LC, Solomon LM, Zeis PM, Justice P, Rosenthal IM. Vitamin D-resistant rickets associated with epidermal nevus syndrome: demonstration of a phosphaturic substance in the dermal lesions. J Pediatr. 1977;91:56–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR051598/AR/NIAMS NIH HHS/United States
- R03 DE015900/DE/NIDCR NIH HHS/United States
- 1 R01 AR51598-01/AR/NIAMS NIH HHS/United States
- 1 R03 DE015900-01/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous