Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine - PubMed (original) (raw)
Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine
Yuxian He et al. Biochem Biophys Res Commun. 2004.
Abstract
The spike (S) protein of severe acute respiratory syndrome (SARS) coronavirus (CoV), a type I transmembrane envelope glycoprotein, consists of S1 and S2 domains responsible for virus binding and fusion, respectively. The S1 contains a receptor-binding domain (RBD) that can specifically bind to angiotensin-converting enzyme 2 (ACE2), the receptor on target cells. Here we show that a recombinant fusion protein (designated RBD-Fc) containing 193-amino acid RBD (residues 318-510) and a human IgG1 Fc fragment can induce highly potent antibody responses in the immunized rabbits. The antibodies recognized RBD on S1 domain and completely inhibited SARS-CoV infection at a serum dilution of 1:10,240. Rabbit antisera effectively blocked binding of S1, which contains RBD, to ACE2. This suggests that RBD can induce highly potent neutralizing antibody responses and has potential to be developed as an effective and safe subunit vaccine for prevention of SARS.
Figures
Fig. 1
Schematic diagram of SAR-CoV S protein and the recombinant fusion protein RBD-Fc. The S protein consists of S1 and S2 domains. There is a signal peptide (SP) located at the N-terminus of the S protein. The S1 domain contains a receptor-binding domain (RBD). The S2 domain contains a cytoplasm domain (CP), a transmembrane domain (TM), and an ectodomain composed of a putative internal fusion peptide (FP) and heptad repeat 1 and 2 (HR1 and HR2) regions. RBD-Fc consists of RBD and a human IgG-Fc fragment. S1-C9 contains S protein S1 domain and a C9 fragment.
Fig. 2
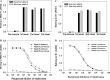
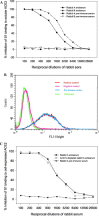
Rabbit antisera contained high titers of antibodies binding to RBD. (A) Binding to RBD-Fc by antisera (1:10,000) collected from rabbits before immunization (pre-immune) and 10 days after each boost; (B) binding to RBD-Fc by rabbit antisera collected 10 days after the first boost at a series of 5-fold dilutions; (C) binding to S1-C9 by antisera (1:10,000) collected from rabbits before immunization (pre-immune) and 10 days after each boost; and (D) binding to S1-C9 protein by rabbit antisera collected 10 days after the first boost at a series of 5-fold dilutions. All samples were tested in duplicate and data presented are mean values of two tests (same for the following figures).
Fig. 3
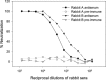
Neutralization of SARS-CoV by rabbit antisera directed against RBD-Fc. SARS-CoV was incubated with Vero E6 monolayer in the presence of rabbit antisera in a series of 2-fold dilutions. The CPE caused by SARS-CoV infection was recorded under microscope and the virus-neutralizing titers were calculated.
Fig. 4
Neutralization of HIV/SARS-CoV S pseudovirus infection by rabbit antisera. Inhibition of a single-cycle infection of 293T cells expressing ACE2 by the pseudovirus was determined in a luciferase assay.
Fig. 5
Effect of depletion of human IgG-Fc specific antibodies from the rabbit antisera on binding to S1 and virus-neutralizing activity. The binding activity of anti-Fc-depleted and untreated rabbit antisera to human IgG (A) and S1 (B) was tested at 1:50 dilution by ELISA. The neutralizing activity of the anti-Fc-depleted rabbit antisera against HIV/SARS-CoV S was compared with that of untreated rabbit antisera (C).
Fig. 6
Rabbit antisera inhibited S1 binding to ACE2. (A) inhibition of S1 binding to soluble ACE2 by rabbit antisera was measured by ELISA; (B) inhibition of S1 binding to cell-expressed ACE2 by rabbit antisera was measured by flow cytometry. In the positive control, no rabbit serum was added while in the negative control, neither rabbit serum nor S1-C9 was added; (C) rabbit antisera inhibited S1 binding to ACE2-expressing cells in a dose-dependent manner.
Similar articles
- A single amino acid substitution (R441A) in the receptor-binding domain of SARS coronavirus spike protein disrupts the antigenic structure and binding activity.
He Y, Li J, Jiang S. He Y, et al. Biochem Biophys Res Commun. 2006 May 26;344(1):106-13. doi: 10.1016/j.bbrc.2006.03.139. Epub 2006 Mar 30. Biochem Biophys Res Commun. 2006. PMID: 16615996 Free PMC article. - Recombinant Receptor-Binding Domains of Multiple Middle East Respiratory Syndrome Coronaviruses (MERS-CoVs) Induce Cross-Neutralizing Antibodies against Divergent Human and Camel MERS-CoVs and Antibody Escape Mutants.
Tai W, Wang Y, Fett CA, Zhao G, Li F, Perlman S, Jiang S, Zhou Y, Du L. Tai W, et al. J Virol. 2016 Dec 16;91(1):e01651-16. doi: 10.1128/JVI.01651-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795425 Free PMC article. - Receptor-binding domain of SARS-Cov spike protein: soluble expression in E. coli, purification and functional characterization.
Chen J, Miao L, Li JM, Li YY, Zhu QY, Zhou CL, Fang HQ, Chen HP. Chen J, et al. World J Gastroenterol. 2005 Oct 21;11(39):6159-64. doi: 10.3748/wjg.v11.i39.6159. World J Gastroenterol. 2005. PMID: 16273643 Free PMC article. - Perspectives on the development of neutralizing antibodies against SARS-CoV-2.
Ho M. Ho M. Antib Ther. 2020 Apr;3(2):109-114. doi: 10.1093/abt/tbaa009. Epub 2020 May 20. Antib Ther. 2020. PMID: 32566896 Free PMC article. Review.
Cited by
- Virus pathogen database and analysis resource (ViPR): a comprehensive bioinformatics database and analysis resource for the coronavirus research community.
Pickett BE, Greer DS, Zhang Y, Stewart L, Zhou L, Sun G, Gu Z, Kumar S, Zaremba S, Larsen CN, Jen W, Klem EB, Scheuermann RH. Pickett BE, et al. Viruses. 2012 Nov 19;4(11):3209-26. doi: 10.3390/v4113209. Viruses. 2012. PMID: 23202522 Free PMC article. - Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates.
Walsh EE, Frenck RW Jr, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi PY, Türeci Ö, Tompkins KR, Lyke KE, Raabe V, Dormitzer PR, Jansen KU, Şahin U, Gruber WC. Walsh EE, et al. N Engl J Med. 2020 Dec 17;383(25):2439-2450. doi: 10.1056/NEJMoa2027906. Epub 2020 Oct 14. N Engl J Med. 2020. PMID: 33053279 Free PMC article. Clinical Trial. - Landscape and selection of vaccine epitopes in SARS-CoV-2.
Smith CC, Olsen KS, Gentry KM, Sambade M, Beck W, Garness J, Entwistle S, Willis C, Vensko S, Woods A, Fini M, Carpenter B, Routh E, Kodysh J, O'Donnell T, Haber C, Heiss K, Stadler V, Garrison E, Sandor AM, Ting JPY, Weiss J, Krajewski K, Grant OC, Woods RJ, Heise M, Vincent BG, Rubinsteyn A. Smith CC, et al. Genome Med. 2021 Jun 14;13(1):101. doi: 10.1186/s13073-021-00910-1. Genome Med. 2021. PMID: 34127050 Free PMC article. - Induction of Potent and Durable Neutralizing Antibodies Against SARS-CoV-2 Using a Receptor Binding Domain-Based Immunogen.
Srivastava V, Niu L, Phadke KS, Bellaire BH, Cho MW. Srivastava V, et al. Front Immunol. 2021 Mar 11;12:637982. doi: 10.3389/fimmu.2021.637982. eCollection 2021. Front Immunol. 2021. PMID: 33777030 Free PMC article. - Recent Advancements in the Diagnosis, Prevention, and Prospective Drug Therapy of COVID-19.
Ahsan W, Alhazmi HA, Patel KS, Mangla B, Al Bratty M, Javed S, Najmi A, Sultan MH, Makeen HA, Khalid A, Mohan S, Taha MME, Sultana S. Ahsan W, et al. Front Public Health. 2020 Jul 10;8:384. doi: 10.3389/fpubh.2020.00384. eCollection 2020. Front Public Health. 2020. PMID: 32754570 Free PMC article. Review.
References
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
- Drosten C., Gunther S., Preiser W., Van Der W.S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
- Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. - PubMed
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous