A high resolution protein interaction map of the yeast Mediator complex - PubMed (original) (raw)
. 2004 Oct 11;32(18):5379-91.
doi: 10.1093/nar/gkh878. Print 2004.
Affiliations
- PMID: 15477388
- PMCID: PMC524289
- DOI: 10.1093/nar/gkh878
A high resolution protein interaction map of the yeast Mediator complex
Benjamin Guglielmi et al. Nucleic Acids Res. 2004.
Abstract
Mediator is a large, modular protein complex remotely conserved from yeast to man that conveys regulatory signals from DNA-binding transcription factors to RNA polymerase II. In Saccharomyces cerevisiae, Mediator is thought to be composed of 24 subunits organized in four sub-complexes, termed the head, middle, tail and Cdk8 (Srb8-11) modules. In this work, we have used screening and pair-wise two-hybrid approaches to investigate protein-protein contacts between budding yeast Mediator subunits. The derived interaction map includes the delineation of numerous interaction domains between Mediator subunits, frequently corresponding to segments that have been conserved in evolution, as well as novel connections between the Cdk8 (Srb8-11) and head modules, the head and middle modules, and the middle and tail modules. The two-hybrid analysis, together with co-immunoprecipitation studies and gel filtration experiments revealed that Med31 (Soh1) is associated with the yeast Mediator that therefore comprises 25 subunits. Finally, analysis of the protein interaction network within the Drosophila Mediator middle module indicated that the structural organization of the Mediator complex is conserved from yeast to metazoans. The resulting interaction map provides a framework for delineating Mediator structure-function and investigating how Mediator function is regulated.
Figures
Figure 1
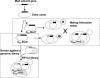
Experimental scheme. Yeast Mediator subunit genes were individually cloned into a Gateway entry vector. Each Mediator ORF was then fused to the Gal4 DNA-binding (GBD–MedX) or activation domain (GAD–MedY). Each GBD–MedX fusion protein was tested for two-hybrid interactions in a mating assay against each GAD–MedY fusion protein. The GBD–MedX fusion proteins were also screened against the FRYL2 library of yeast genomic fragments fused to the GAD domain. _GUAS_-lacZ and _GUAS_-HIS3 represent the GAL1::lacZ and GAL4(UAS)::HIS3 reporter genes, respectively.
Figure 2
Mating assay for Mediator subunit interactions. An example of the mating assay is shown where Y190 strains transformed by plasmids encoding fusions of the GBD with Med12 (Srb8), Med17 (Srb4), Med18 (Srb5), Med21 (Srb7), Med22 (Srb6) or Cdk8 (Srb10) [Med13 (Srb9) was omitted since it activated the transcription of the reporter gene] were crossed with Y187 derivatives transformed with plasmids encoding fusions of the GAD with Med11, Med12 (Srb8), Med13 (Srb9), Med17 (Srb4), Med18 (Srb5), Med20 (Srb2), Med21 (Srb7) or Med22 (Srb6). Patches of cells were overlaid with X-Gal agarose (31) to reveal β-galactosidase activity (blue colour). The interactions between Med11 and Med17 (Srb4), Med11 and Med22 (Srb6), Med18 (Srb5) and Med20 (Srb2), as well as between Med17 (Srb4) and Med22 (Srb6) can be observed. The mating control indicated that haploid parents did not grow. A contamination control was included to detect carry over from separate samples in the 96 well plates.
Figure 3
Two-hybrid interactions between yeast Mediator subunits. The two-hybrid interactions between the yeast Mediator subunits are indicated on the left-hand side half-matrix. The interactions that were only found in two-hybrid screens with the FRYL2 library are indicated by an upward pointing closed triangle, those that were only observed in the mating assays are indicated by an open circle, and those that were found in both assays are indicated by a closed circle. The asterisk on Med31 (Soh1) indicates that it was only tested as a GBD fusion after it was found in the Med10 and Med21 (Srb7) screens, since Med31 (Soh1) was not taken into account initially because it was not previously considered as a yeast Mediator subunit. Results from previously published screens are indicated in the upper right half of the figure. Black squares represent GST pull-down results (–26), downward pointing open triangles indicate two-hybrid interactions from Uetz et al. (27), open diamonds those from Ito et al. (28), black triangles interactions observed both by GST pull-down and Uetz et al. (27), and black diamonds interactions observed by GST pull-down and Ito et al. (28).
Figure 4
Med31 (Soh1) is associated with yeast Mediator. (A) Med31 (Soh1) two-hybrid interaction with Med10. Patches of cells were treated as in Figure 2. GAD and GBD indicate empty vectors used as negative controls. GBD–Med7 was used as a positive control for Med10 interaction. (B) Med31 (Soh1) and Med17 (Srb4) co-immunoprecipitate. After immunoprecipitation, the proteins were revealed by western blotting using 12CA5 or PAP antibodies, binding to the HA or TAP tag, respectively. Top panel: anti-HA IP; bottom panel: PAP IP. Negative controls were strains expressing Med17–TAP (Srb4–TAP) or Med31–HA (Soh1–HA) only. (C) Gel filtration analysis of extracts from strains expressing Med31 (Soh1) and Med17 (Srb4) after anti-HA immunoprecipitation. The immunoprecipitation extract was run on a Superose 6 gel filtration column calibrated with molecular weight markers. The presence of Med31–HA (Soh1–HA) or Med17–TAP (Srb4–TAP) was revealed by western blotting as in (B). The amount of protein was measured by densitometric analyses of films and is expressed in arbitrary units on the left scale. Measuring UV absorbance during chromatography (right scale) assessed the position of the markers. The dashed line indicates the UV trace. The molecular weight of the markers is indicated above their respective peak abundance.
Figure 5
Analysis of direct interactions of yeast Med4, Med7 and Med8 with their partners. Med4 protein fragments used to analyse the interactions of Med4, Med7 and Med8 with their partners are represented. + indicates GAL1::lacZ activation; − indicates absence of activation. The Mediator subunit conserved domains defined according to Boube et al. (10) are indicated by black boxes. The conservation of the C-terminal domain of Med8 was not reported previously. An alignment is shown in supplementary Figure S1.
Figure 6
Interaction domains map to conserved regions of the Mediator subunits. The conserved domains of the Mediator proteins for which interaction domains could be mapped are represented by black boxes. The solid lines indicate the extent of the interaction domains (10) (Figure S1). The number of independent clones selected in the screens is indicated between parenthesis.
Figure 7
Interactions between the D.melanogaster Mediator middle module subunits The two-hybrid interactions between the predicted D.melanogaster Mediator middle module subunits are indicated in the half-matrix. CG5134 was predicted by Tomomori-Sato et al. (49) to be homologous to Med9. The other homology predictions are those of Boube et al. (10). The activation of the GAL1::lacZ reporter gene to reveal the interaction between Med7 and Med21 (Srb7) required cultivation of the transformed strain on 3-AT medium.
Figure 8
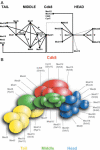
Integrated interaction map of the yeast Mediator. (A) Connection map of Mediator subunits. The direct links between the different Mediator subunits found in this work, by Kang et al. (–26), Ito et al. (28) and Uetz et al. (27) are combined to produce the integrated Mediator map. The interactions between Med17 (Srb4) and Med19 (Rox3) or Med20 (Srb2) (24) and between Med6 and Med21 (38) that were not found or confirmed in our screens or mating assays are indicated by grey lines. Black stars indicate the interactions that were found with the Mediator middle module homologous subunits from Drosophila. The grey star indicates a conserved contact between the worm Med22 (MDT-22/ZK970.3) and Med11 (MDT-11/R144.9) homologues (44). (B) Topological organization of yeast Mediator. This model was made taking into account all data mentioned in (A) and the relative size of the Mediator subunits.
Similar articles
- Structure of the Mediator head module.
Larivière L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P. Larivière L, et al. Nature. 2012 Dec 20;492(7429):448-51. doi: 10.1038/nature11670. Epub 2012 Oct 31. Nature. 2012. PMID: 23123849 - The Soh1/MED31 protein is an ancient component of Schizosaccharomyces pombe and Saccharomyces cerevisiae Mediator.
Linder T, Gustafsson CM. Linder T, et al. J Biol Chem. 2004 Nov 19;279(47):49455-9. doi: 10.1074/jbc.M409046200. Epub 2004 Sep 8. J Biol Chem. 2004. PMID: 15356001 - Tail and Kinase Modules Differently Regulate Core Mediator Recruitment and Function In Vivo.
Jeronimo C, Langelier MF, Bataille AR, Pascal JM, Pugh BF, Robert F. Jeronimo C, et al. Mol Cell. 2016 Nov 3;64(3):455-466. doi: 10.1016/j.molcel.2016.09.002. Epub 2016 Oct 20. Mol Cell. 2016. PMID: 27773677 Free PMC article. - The yeast Mediator complex and its regulation.
Björklund S, Gustafsson CM. Björklund S, et al. Trends Biochem Sci. 2005 May;30(5):240-4. doi: 10.1016/j.tibs.2005.03.008. Trends Biochem Sci. 2005. PMID: 15896741 Review. - Twenty years of Mediator complex structural studies.
Verger A, Monté D, Villeret V. Verger A, et al. Biochem Soc Trans. 2019 Feb 28;47(1):399-410. doi: 10.1042/BST20180608. Epub 2019 Feb 7. Biochem Soc Trans. 2019. PMID: 30733343 Free PMC article. Review.
Cited by
- The Mediator complex: a central integrator of transcription.
Allen BL, Taatjes DJ. Allen BL, et al. Nat Rev Mol Cell Biol. 2015 Mar;16(3):155-66. doi: 10.1038/nrm3951. Epub 2015 Feb 18. Nat Rev Mol Cell Biol. 2015. PMID: 25693131 Free PMC article. Review. - Functional and physical interactions within the middle domain of the yeast mediator.
Hallberg M, Hu GZ, Tronnersjö S, Adler D, Balciunas D, Björklund S, Ronne H. Hallberg M, et al. Mol Genet Genomics. 2006 Aug;276(2):197-210. doi: 10.1007/s00438-006-0135-7. Epub 2006 Jun 7. Mol Genet Genomics. 2006. PMID: 16758199 - Activator Gcn4 employs multiple segments of Med15/Gal11, including the KIX domain, to recruit mediator to target genes in vivo.
Jedidi I, Zhang F, Qiu H, Stahl SJ, Palmer I, Kaufman JD, Nadaud PS, Mukherjee S, Wingfield PT, Jaroniec CP, Hinnebusch AG. Jedidi I, et al. J Biol Chem. 2010 Jan 22;285(4):2438-55. doi: 10.1074/jbc.M109.071589. Epub 2009 Nov 23. J Biol Chem. 2010. PMID: 19940160 Free PMC article. - Distinct roles for Mediator Cdk8 module subunits in Drosophila development.
Loncle N, Boube M, Joulia L, Boschiero C, Werner M, Cribbs DL, Bourbon HM. Loncle N, et al. EMBO J. 2007 Feb 21;26(4):1045-54. doi: 10.1038/sj.emboj.7601566. Epub 2007 Feb 8. EMBO J. 2007. PMID: 17290221 Free PMC article.
References
- Kim Y.-J., Björklund,S., Li,Y., Sayre,M.H. and Kornberg,R.D. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell, 77, 599–608. - PubMed
- Thompson C.M., Koleske,A.J., Chao,D.M. and Young,R.A. (1993) A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell, 73, 1361–1375. - PubMed
- Holstege F.C.P., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. - PubMed
- Ito M., Yuan,C.X., Malik,S., Gu,W., Fondell,J.D., Yamamura,S., Fu,Z.Y., Zhang,X., Qin,J. and Roeder,R.G. (1999) Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell, 3, 361–370. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases