Tumor necrosis factor alpha induction of NF-kappaB requires the novel coactivator SIMPL - PubMed (original) (raw)
Tumor necrosis factor alpha induction of NF-kappaB requires the novel coactivator SIMPL
Hyung-Joo Kwon et al. Mol Cell Biol. 2004 Nov.
Abstract
A myriad of stimuli including proinflammatory cytokines, viruses, and chemical and mechanical insults activate a kinase complex composed of IkappaB kinase beta (IKK-beta), IKK-alpha, and IKK-gamma/N, leading to changes in NF-kappaB-dependent gene expression. However, it is not clear how the NF-kappaB response is tailored to specific cellular insults. Signaling molecule that interacts with mouse pelle-like kinase (SIMPL) is a signaling component required for tumor necrosis factor alpha (TNF-alpha)-dependent but not interleukin-1-dependent NF-kappaB activation. Herein we demonstrate that nuclear localization of SIMPL is required for type I TNF receptor-induced NF-kappaB activity. SIMPL interacts with nuclear p65 in a TNF-alpha-dependent manner to promote endogenous NF-kappaB-dependent gene expression. The interaction between SIMPL and p65 enhances p65 transactivation activity. These data support a model in which TNF-alpha activation of NF-kappaB dependent-gene expression requires nuclear relocalization of p65 as well as nuclear relocalization of SIMPL, generating a TNF-alpha-specific induction of gene expression.
Figures
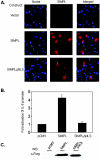
FIG.1.
SIMPL contains a nuclear localization signal that is required for activation of NF-κB. (A) HEK 293 cells were transfected with vector alone or a vector encoding Flag-tagged SIMPL or Flag-tagged SIMPLΔNLS. Twenty-four hours later, cultures were fixed, processed for indirect immunofluorescence with Flag-specific antisera (column labeled SIMPL), or stained with Hoechst no. 33258 to visualize the nuclei (column labeled Nuclei). (B and C) HEK 293 cells were transfected with an IL-8 promoter-firefly luciferase construct, a promoterless sea pansy luciferase construct, and 0.5 μg of the indicated plasmid constructs. Forty-eight hours later, cultures were harvested and either (B) subjected to dual-luciferase assays as described under Materials and Methods or (C) normalized for protein content and subjected to Western analysis using the indicated antisera.
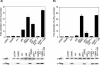
FIG. 2.
Nuclear localization of SIMPL is required for TNF RI-dependent NF-κB activation. (A and B) HEK 293 cells were transfected with 0.5 μg of the indicated constructs, 20 ng of the promoterless sea pansy luciferase construct, and 0.2 μg of either (A) a firefly luciferase construct under the control of the IL-8 gene promoter or (B) an artificial promoter composed of three tandem repeats of the NF-κB response element from the human immunodeficiency virus long terminal repeat. Twenty-four hours later, cultures were harvested and processed for luciferase activity as described in Materials and Methods.
FIG. 3.
SIMPL modulates p65 and not c-Jun- or C/EBP-dependent transcription. (A to D) HEK 293 cells were transfected with 0.5 μg of the indicated constructs, 20 ng of the promoterless sea pansy luciferase construct, and 0.2 μg of either (A and C) a firefly luciferase construct under the control of the IL-8 gene promoter or (B and D) an artificial promoter composed of three tandem repeats of the NF-κB response element from the human immunodeficiency virus long terminal repeat. Twenty-four hours later, cultures were harvested and processed for luciferase activity as described in Materials and Methods. In all experiments, cell lysates normalized for protein content were subjected to Western analysis to confirm expression of the Flag-tagged SIMPL construct and the p65 construct. (E) HEK cells were transfected with 0.5 μg of the indicated constructs. Twenty-four hours later, cultures were harvested, RNA was isolated, and RNase protection assays were performed as described in Materials and Methods. (F) The autoradiogram used to generate the figure in panel E was quantitated by densitometry, and the ratio of the values obtained (IL-8/GAPDH) was replotted as a histogram.
FIG. 3.
SIMPL modulates p65 and not c-Jun- or C/EBP-dependent transcription. (A to D) HEK 293 cells were transfected with 0.5 μg of the indicated constructs, 20 ng of the promoterless sea pansy luciferase construct, and 0.2 μg of either (A and C) a firefly luciferase construct under the control of the IL-8 gene promoter or (B and D) an artificial promoter composed of three tandem repeats of the NF-κB response element from the human immunodeficiency virus long terminal repeat. Twenty-four hours later, cultures were harvested and processed for luciferase activity as described in Materials and Methods. In all experiments, cell lysates normalized for protein content were subjected to Western analysis to confirm expression of the Flag-tagged SIMPL construct and the p65 construct. (E) HEK cells were transfected with 0.5 μg of the indicated constructs. Twenty-four hours later, cultures were harvested, RNA was isolated, and RNase protection assays were performed as described in Materials and Methods. (F) The autoradiogram used to generate the figure in panel E was quantitated by densitometry, and the ratio of the values obtained (IL-8/GAPDH) was replotted as a histogram.
FIG. 3.
SIMPL modulates p65 and not c-Jun- or C/EBP-dependent transcription. (A to D) HEK 293 cells were transfected with 0.5 μg of the indicated constructs, 20 ng of the promoterless sea pansy luciferase construct, and 0.2 μg of either (A and C) a firefly luciferase construct under the control of the IL-8 gene promoter or (B and D) an artificial promoter composed of three tandem repeats of the NF-κB response element from the human immunodeficiency virus long terminal repeat. Twenty-four hours later, cultures were harvested and processed for luciferase activity as described in Materials and Methods. In all experiments, cell lysates normalized for protein content were subjected to Western analysis to confirm expression of the Flag-tagged SIMPL construct and the p65 construct. (E) HEK cells were transfected with 0.5 μg of the indicated constructs. Twenty-four hours later, cultures were harvested, RNA was isolated, and RNase protection assays were performed as described in Materials and Methods. (F) The autoradiogram used to generate the figure in panel E was quantitated by densitometry, and the ratio of the values obtained (IL-8/GAPDH) was replotted as a histogram.
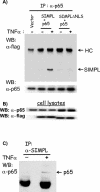
FIG. 4.
SIMPL-p65 complexes form in response to TNF-α. (A and B) HEK 293 cells were transfected with an expression vector encoding p65 and an expression vector encoding either wild-type SIMPL or SIMPLΔNLS. Twenty-four hours later, the indicated cultures were treated with recombinant human TNF-α and were harvested 15 min later. Cell lysates were prepared, and p65 antisera were used to generate immunocomplexes. (A) Immunocomplexed materials or (B) cell lysates used to generate the immunocomplexes were analyzed by Western analysis for SIMPL or p65. (C) Duplicate sets of HEK 293 cell cultures were plated; 24 h later, one set was not treated and the second set was treated with rhuTNFα for 15 min; all cultures were then harvested. Cell lysates were prepared, and immunocomplexes were generated with SIMPL antisera. Immunocomplexes (α-SIMPL) were subjected to SDS-PAGE, and Western blots were prepared and probed with p65 antisera.
FIG. 5.
Loss of SIMPL does not prevent nuclear localization of p65. (A) Duplicate sets of C3H10T1/2 mouse embryo fibroblasts were transfected with 0.5 μg of the indicated constructs. Twenty-four hours later, half of the cultures were treated with TNF-α (+), and all cultures were fixed and washed with PBS 15 min later. Cultures were processed for indirect immunofluorescence by using antiserum that recognizes p65. (B) C3H10T1/Z mouse embryo fibroblasts were transfected with a construct encoding Flag-SIMPL. Forty-eight hours later, cultures were treated with recombinant human TNF-α (10 ng/ml) for 15 min. Cytoplasmic and nuclear fractions were prepared, and equal amounts of cellular protein were subjected to SDS-PAGE. A Western blot was prepared and probed with p65 or Flag-specific antiserum.
FIG. 6.
SIMPL functions as a p65 coactivator. (A) HEK 293 cells were transfected with 1.0 μg of the indicated constructs, 5 ng of a p65-Gal4 construct, and 200 ng of a Gal4-luciferase construct. Cultures were allowed to incubate for 24 to 36 h, at which time cells were harvested and lysates were made and assayed for luciferase activity. (B) Wild-type (first three bars) or RelA−/− (last three bars) MEFs were transfected with 250 ng of the indicated vectors plus 200 ng of an IL-8-luciferase construct and 20 ng of the promoterless sea pansy luciferase construct. Twenty-four hours later, cultures were harvested and luciferase activities were measured. (C) HEK 293 cells were transfected with empty vector (control) or a construct expressing antisense SIMPL. Twenty-four hours later, cultures were stimulated with recombinant human-TNF-α (10 ng/ml), and cultures were harvested at the indicated times. Cell lysates, normalized by protein concentration, were subjected to SDS-PAGE, Western blots were prepared and probed with IκBα and GAPDH antisera. Films were scanned and quantitated by using TotalLab (Nonlinear Dynamics). (D) Model of TNF control of NF-κB-dependent gene expression.
References
- Chen, Z. J., L. Parent, and T. Maniatis. 1996. Site-specific phosphorylation of IkBa by a novel ubiquitination-dependent protein kinase activity. Cell 84:853-862. - PubMed
- DiDonato, J. A., M. Hayakawa, D. M. Rothwarf, E. Zandi, and M. Karin. 1997. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature 388:548-554. - PubMed
- Efthymiadis, A., H. Shao, S. Hubner, and D. A. Jans. 1997. Kinetic characterization of the human retinoblastoma protein bipartite nuclear localization sequence (NLS) in vivo and in vitro. J. Biol. Chem. 272:22134-22139. - PubMed
- Fontes, M. R. M., T. The, and B. Kobe. 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol. 297:1183-1194. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases