High-throughput expression of C. elegans proteins - PubMed (original) (raw)
. 2004 Oct;14(10B):2102-10.
doi: 10.1101/gr.2520504.
Shihong Qiu, James B Finley, Mike Carson, Rita J Gray, Wenying Huang, David Johnson, Jun Tsao, Jérôme Reboul, Philippe Vaglio, David E Hill, Marc Vidal, Lawrence J Delucas, Ming Luo
Affiliations
- PMID: 15489332
- PMCID: PMC528926
- DOI: 10.1101/gr.2520504
High-throughput expression of C. elegans proteins
Chi-Hao Luan et al. Genome Res. 2004 Oct.
Abstract
Proteome-scale studies of protein three-dimensional structures should provide valuable information for both investigating basic biology and developing therapeutics. Critical for these endeavors is the expression of recombinant proteins. We selected Caenorhabditis elegans as our model organism in a structural proteomics initiative because of the high quality of its genome sequence and the availability of its ORFeome, protein-encoding open reading frames (ORFs), in a flexible recombinational cloning format. We developed a robotic pipeline for recombinant protein expression, applying the Gateway cloning/expression technology and utilizing a stepwise automation strategy on an integrated robotic platform. Using the pipeline, we have carried out heterologous protein expression experiments on 10,167 ORFs of C. elegans. With one expression vector and one Escherichia coli strain, protein expression was observed for 4854 ORFs, and 1536 were soluble. Bioinformatics analysis of the data indicates that protein hydrophobicity is a key determining factor for an ORF to yield a soluble expression product. This protein expression effort has investigated the largest number of genes in any organism to date. The pipeline described here is applicable to high-throughput expression of recombinant proteins for other species, both prokaryotic and eukaryotic, provided that ORFeome resources become available.
Figures
Figure 1
Reproducibility of protein expression in 96-well format. The data are from two experiments of protein expression, purification, and solubility profiling for plate 19. The data demonstrate good reproducibility for identifying soluble proteins in the multigene, multistep experiments. The data on left in 96-well format are ELISA readings of OD (optical density) at 405 nm for the supernatant (soluble fraction) of the expression, each well for one ORF. The maximum OD reading is 4.0. Color coding describes the level of protein expression, where red is for high, orange for medium, and yellow for low. On the right, listed with OD for each soluble protein, is σ which is calculated as the ratio of OD reading for the supernatant (soluble fraction, data shown) to that for pellet (insoluble fraction, data not shown) from the same expression experiment of an ORF. The OD and σ values combined serve as the solubility signature of a protein.
Figure 2
Temperature dependence of total and soluble protein expression in 96-well format. More soluble proteins were found at 18°C; more proteins were expressed at 37°C. For this plate, the total protein expression was about 40%, whereas the soluble expression was ∼12%. The data are OD readings from 96-well format ELISA for ORFs in plate 51. Color coding is the same as in Figure 1.
Figure 3
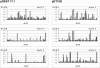
Comparison of expression vector pDEST17.1 vs. pET15G for protein expression. Protein expression of the genes in the four plates was carried out using pDEST17.1 (plots on left) and pET15G (plots on right). The number of soluble proteins was increased in all cases by using pET15G. The data are represented as histograms of OD readings at 405 nm from ELISA vs. ORFs in the 96-well plate.
Figure 4
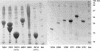
SDS-polyacrylamide gels showing the 1-L scale-up expressions of 12 soluble proteins. The expression levels in the gel are typical for the C. elegans proteins expressed in E. coli. The bands for 76D4, 76F6, 56D11, and 76F10 represents a high-level expression with yield greater than 10 mg per liter of bacterial cell culture (mg/L). The bands for 37F1, 37G9, and 37C1 represent a medium level expression from 6 to 10 mg/L, whereas the bands for 37D4, 37B8, and 37D8 represent a low level expression from 3 to 6 mg/L.
Figure 5
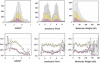
Histograms (top) of GRAVY, molecular weight, and isoelectric point for the 10,167 ORFs studied (white), expressed (yellow), and soluble (pink), and percentage plots (bottom) of the expressed and soluble ORFs over the studied, respectively, vs. the three parameters. Correlation between protein expression and GRAVY is apparent in the plots. The lack of a correlation to isoelectric point and molecular weight is indicated by the flatness of the curves in the percentage plots.
Figure 6
Expression data in correlation to GRAVY, Signal peptide, and Transmembrane helices as demonstrated on 87 ORFs for plate 11041. The genes are listed in two panels ordered by GRAVY value from low to high in the left panel and continued in the same manner in the right panel. The break point for the two panels was chosen for easy presentation. (Right) Higher GRAVY values. The expression data are color coded with gray for no expression, white for expression but not soluble, yellow for low level soluble, orange for mid level soluble, and red for high level soluble. (SP) Signal peptide or anchor; (TM) number of transmembrane helices.
Figure 7
Schematic for the HTP protein expression pipeline. The rectangles represent material, the arrows represent process, and the octagon represents data.
Similar articles
- A high throughput platform for eukaryotic genes.
Chen Y, Qiu S, Luan CH, Luo M. Chen Y, et al. Methods Mol Biol. 2008;426:209-20. doi: 10.1007/978-1-60327-058-8_13. Methods Mol Biol. 2008. PMID: 18542866 - Domain selection combined with improved cloning strategy for high throughput expression of higher eukaryotic proteins.
Chen Y, Qiu S, Luan CH, Luo M. Chen Y, et al. BMC Biotechnol. 2007 Jul 30;7:45. doi: 10.1186/1472-6750-7-45. BMC Biotechnol. 2007. PMID: 17663785 Free PMC article. - C. elegans ORFeome version 3.1: increasing the coverage of ORFeome resources with improved gene predictions.
Lamesch P, Milstein S, Hao T, Rosenberg J, Li N, Sequerra R, Bosak S, Doucette-Stamm L, Vandenhaute J, Hill DE, Vidal M. Lamesch P, et al. Genome Res. 2004 Oct;14(10B):2064-9. doi: 10.1101/gr.2496804. Genome Res. 2004. PMID: 15489327 Free PMC article. - ORFeome projects: gateway between genomics and omics.
Rual JF, Hill DE, Vidal M. Rual JF, et al. Curr Opin Chem Biol. 2004 Feb;8(1):20-5. doi: 10.1016/j.cbpa.2003.12.002. Curr Opin Chem Biol. 2004. PMID: 15036152 Review. - Many paths to many clones: a comparative look at high-throughput cloning methods.
Marsischky G, LaBaer J. Marsischky G, et al. Genome Res. 2004 Oct;14(10B):2020-8. doi: 10.1101/gr.2528804. Genome Res. 2004. PMID: 15489321 Review.
Cited by
- Prediction of Solubility of Proteins in Escherichia coli Based on Functional and Structural Features Using Machine Learning Methods.
Huang F, Gao Q, Zhou X, Guo W, Feng K, Zhu L, Huang T, Cai YD. Huang F, et al. Protein J. 2024 Oct;43(5):983-996. doi: 10.1007/s10930-024-10230-z. Epub 2024 Sep 7. Protein J. 2024. PMID: 39243320 - Massively parallel interrogation of protein fragment secretability using SECRiFY reveals features influencing secretory system transit.
Boone M, Ramasamy P, Zuallaert J, Bouwmeester R, Van Moer B, Maddelein D, Turan D, Hulstaert N, Eeckhaut H, Vandermarliere E, Martens L, Degroeve S, De Neve W, Vranken W, Callewaert N. Boone M, et al. Nat Commun. 2021 Nov 5;12(1):6414. doi: 10.1038/s41467-021-26720-y. Nat Commun. 2021. PMID: 34741024 Free PMC article. - Developments and Applications of Functional Protein Microarrays.
Syu GD, Dunn J, Zhu H. Syu GD, et al. Mol Cell Proteomics. 2020 Jun;19(6):916-927. doi: 10.1074/mcp.R120.001936. Epub 2020 Apr 17. Mol Cell Proteomics. 2020. PMID: 32303587 Free PMC article. Review. - Correlation Between Protein Primary Structure and Soluble Expression Level of HSA dAb in Escherichia coli.
Yang Y, Liu G, Liu M, Bai Z, Liu X, Dai X, Guo W. Yang Y, et al. Food Technol Biotechnol. 2018 Mar;56(1):101-109. doi: 10.17113/ftb.56.01.18.5445. Food Technol Biotechnol. 2018. PMID: 29796003 Free PMC article. - Rational identification of aggregation hotspots based on secondary structure and amino acid hydrophobicity.
Matsui D, Nakano S, Dadashipour M, Asano Y. Matsui D, et al. Sci Rep. 2017 Aug 25;7(1):9558. doi: 10.1038/s41598-017-09749-2. Sci Rep. 2017. PMID: 28842596 Free PMC article.
References
- Adams, M.W.W., Dailey, H.A., DeLucas, L.J., Luo, M., Prestegard, J.H., Rose, J.P., and Wang, B.C. 2003. The Southeast collaboratory for structural genomics: A high-throughput gene to structure factory. Acc. Chem. Res. 36: 191-198. - PubMed
- Braun, P. and LaBaer, J. 2003. High throughput protein production for functional proteomics. Trends Biotechnol. 21: 383-388. - PubMed
WEB SITE REFERENCES
- http://sgce.cbse.uab.edu; Structural Genomics of C. elegans.
- http://sgi.com; MineSet decision tree software.
- http://ww.invitrogen.com; Gateway Cloning and Expression Technologies—Invitrogen.
- http://ww1.qiagen.com; Qiagen.
- http://ww1.novagen.com; Novagen.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources