From ORFeome to biology: a functional genomics pipeline - PubMed (original) (raw)
. 2004 Oct;14(10B):2136-44.
doi: 10.1101/gr.2576704.
Dorit Arlt, Wolfgang Huber, Ruth Wellenreuther, Simone Schleeger, Alexander Mehrle, Stephanie Bechtel, Mamatha Sauermann, Ulrike Korf, Rainer Pepperkok, Holger Sültmann, Annemarie Poustka
Affiliations
- PMID: 15489336
- PMCID: PMC528930
- DOI: 10.1101/gr.2576704
From ORFeome to biology: a functional genomics pipeline
Stefan Wiemann et al. Genome Res. 2004 Oct.
Abstract
As several model genomes have been sequenced, the elucidation of protein function is the next challenge toward the understanding of biological processes in health and disease. We have generated a human ORFeome resource and established a functional genomics and proteomics analysis pipeline to address the major topics in the post-genome-sequencing era: the identification of human genes and splice forms, and the determination of protein localization, activity, and interaction. Combined with the understanding of when and where gene products are expressed in normal and diseased conditions, we create information that is essential for understanding the interplay of genes and proteins in the complex biological network. We have implemented bioinformatics tools and databases that are suitable to store, analyze, and integrate the different types of data from high-throughput experiments and to include further annotation that is based on external information. All information is presented in a Web database (http://www.dkfz.de/LIFEdb). It is exploited for the identification of disease-relevant genes and proteins for diagnosis and therapy.
Figures
Figure 1
The functional genomics and proteomics pipeline. Starting with the large-scale production and molecular analysis of cDNAs, a human ORFeome resource is generated. This physical resource is systematically exploited in high-throughput applications of protein localization, cell-based assays, and proteomics applications. Information that is derived from these experiments is integrated with expression profiling data from clinical studies and external information to allow for an efficient mining of data. The output is functionally characterized genes and proteins with their possible disease relations. The results are presented through
.
Figure 2
UCSC genome browser view of the gene locus of PRO0971. The exons (numbered bars) and introns (connecting lines) are immediately apparent when cDNAs are aligned with the genome sequence. Arrow heads in the intron lines indicate the orientation of the gene (left to right), with a CpG island (green bar, “CpG: 106”) supporting the 5′-end of the gene and transcript. Multiple coverage of the gene with individual cDNAs (accession nos. BC009485 from the MGC, AK094126 from the FLJ project, and BX647702 from the German cDNA Consortium) helps to identify putative splice variants. An example of exon skipping in the IMAGE:3623656 cDNA (BC009485) as compared with the DKFZp686P0859 cDNA (BX647702) is highlighted within the yellow circle. The UCSC genome browser is at
http://genome.ucsc.edu/cgi-bin/hgGateway
.
Figure 3
Effect of the orientation of the GFP-tag relative to the ORF. (A) For 340 proteins of 567 tested, both orientations resulted in the correct localization of the fusion proteins (same). Another 219 proteins localized differently in the two orientations. Of these, 120 localizations were correctly localizing with the ORF-GFP construct, and 99 fusion proteins localized correctly in the GFP-ORF orientation. Eight expression constructs did not show any detectable expression (none). (B) An example of a mitochondrial protein (upper image). The fusion protein mislocalized (lower image) when the signal peptide at its N terminus was blocked by the GFP-tag. The cytoplasmic and nuclear staining of the GFP-ORF fusion protein is also the default localization of GFP alone. The bar indicates 10 μm.
Figure 4
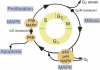
Established assays (gray boxes) to address processes of the cell cycle (yellow circle). G1, S, G2, and M are the phases of the cell cycle (G, growth; S, DNA synthesis; M, mitosis).
Figure 5
Effect of protein overexpression during mitosis. The protein encoded by cDNA DKFZp434P097 was overexpressed as a CFP fusion protein in NIH-3T3 cells (B), antitubulin staining (A). Phosphorylated histone H3 in mitotic cells was detected with a specific antibody (C), which shows a punctuate staining pattern in nuclei of cells in prophase (yellow arrows). The overlay (D) shows colocalization of the DKZp434P097 and the phosphohistone H3 proteins in the cytoplasm. The white arrowhead in _A_-D marks a cell expressing the DKFZp434P097 protein. Bar, 10 μm.
Figure 6
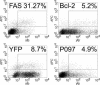
Identification of apoptosis modulators. Shown are plots of the fluorescence intensity in the YFP channel (expression of the recombinant proteins) against the level of activated caspase-3 (measured with an APC-labeled antibody). NIH3T3 cells were transfected with ORFs that were C- or N-terminally tagged with YFP. After 24 h, the cells were stained with an antibody directed against the active form of caspase-3 and measured by FACS. For every protein, the percentage of transfected cells (YFP > 10e1) that were positive for activated caspase-3 (APC > 10e1) is given as compared with the transfected cells (YFP > 10e1) that were negative in active caspase-3 (APC < 10e1). APC is fluorescence of the secondary antibody labeled with allophycocyanine. FAS (Chinnaiyan et al. 1995) is the receptor for the cytokine ligand known as FASL. Activated Fas results in the formation of Death-inducing signaling complex, which ultimately leads to cell death (activator control). Bcl-2 (Hockenbery et al. 1990) is an integral protein of the inner mitochondrial membrane that blocks apoptotic death (inhibitor control). YFP is the YFP protein. P097 is the DKFZp434P097 protein. All proteins were expressed as fusion proteins with YFP.
Figure 7
In vitro phosphorylation of arrayed proteins. Purified proteins were arrayed in quadruplicate on glass slides, and incubated with different protein kinases in the presence of [γ-33P]ATP. Rb is the retinoblastoma protein (Lee et al. 1987), which served as positive control. GFP-GST is purified fusion protein of GFP with a GST-tag, which should not be phosphorylated by the kinases. The protein from cDNA DKFZp434P097 was expressed as a fusion protein with the C terminus of GST. (A) The array was incubated with CDK2/cyclin E kinase. (B) The array was incubated with p42 MAPK kinase.
Figure 8
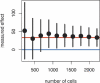
Statistical power analysis for the number of cells. The plot shows means (dots) and 95% confidence intervals (vertical bars) of the measured effect on the proliferation rate of transfection with cyclin A (a positive control in the assay) as a function of the number of cells analyzed. The effect was measured by a robust local regression of the anti-BrdU intensity on the intensity from the YFP-tag (arbitrary fluorescence units). The dependence on the number of cells was simulated by random sampling from the full data set with 2211 cells. The red line represents the approximate true effect, and the blue line no effect. In this example, we would have detected cyclin A as an activator of cell proliferation with 95% probability only for cell numbers ≧1000. Conversely, we would have assigned an activating effect to a protein that is in fact neutral with <5% probability. To reliably detect modifiers of cell proliferation that are subtler, or to achieve probabilities better than 95%, cell numbers must be even higher.
Similar articles
- LIFEdb: a database for functional genomics experiments integrating information from external sources, and serving as a sample tracking system.
Bannasch D, Mehrle A, Glatting KH, Pepperkok R, Poustka A, Wiemann S. Bannasch D, et al. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D505-8. doi: 10.1093/nar/gkh022. Nucleic Acids Res. 2004. PMID: 14681468 Free PMC article. - C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression.
Reboul J, Vaglio P, Rual JF, Lamesch P, Martinez M, Armstrong CM, Li S, Jacotot L, Bertin N, Janky R, Moore T, Hudson JR Jr, Hartley JL, Brasch MA, Vandenhaute J, Boulton S, Endress GA, Jenna S, Chevet E, Papasotiropoulos V, Tolias PP, Ptacek J, Snyder M, Huang R, Chance MR, Lee H, Doucette-Stamm L, Hill DE, Vidal M. Reboul J, et al. Nat Genet. 2003 May;34(1):35-41. doi: 10.1038/ng1140. Nat Genet. 2003. PMID: 12679813 - High-throughput protein analysis integrating bioinformatics and experimental assays.
del Val C, Mehrle A, Falkenhahn M, Seiler M, Glatting KH, Poustka A, Suhai S, Wiemann S. del Val C, et al. Nucleic Acids Res. 2004 Feb 3;32(2):742-8. doi: 10.1093/nar/gkh257. Print 2004. Nucleic Acids Res. 2004. PMID: 14762202 Free PMC article. - Where are we in genomics?
Hocquette JF. Hocquette JF. J Physiol Pharmacol. 2005 Jun;56 Suppl 3:37-70. J Physiol Pharmacol. 2005. PMID: 16077195 Review. - Integration of bioinformatics resources for functional analysis of gene expression and proteomic data.
Huang H, Hu ZZ, Arighi CN, Wu CH. Huang H, et al. Front Biosci. 2007 Sep 1;12:5071-88. doi: 10.2741/2449. Front Biosci. 2007. PMID: 17569631 Review.
Cited by
- Statistical methods and software for the analysis of highthroughput reverse genetic assays using flow cytometry readouts.
Hahne F, Arlt D, Sauermann M, Majety M, Poustka A, Wiemann S, Huber W. Hahne F, et al. Genome Biol. 2006;7(8):R77. doi: 10.1186/gb-2006-7-8-R77. Epub 2006 Aug 17. Genome Biol. 2006. PMID: 16916453 Free PMC article. - Role of the N-terminal activation domain of the coiled-coil coactivator in mediating transcriptional activation by beta-catenin.
Yang CK, Kim JH, Stallcup MR. Yang CK, et al. Mol Endocrinol. 2006 Dec;20(12):3251-62. doi: 10.1210/me.2006-0200. Epub 2006 Aug 24. Mol Endocrinol. 2006. PMID: 16931570 Free PMC article. - Recent advances on host plants and expression cassettes' structure and function in plant molecular pharming.
Makhzoum A, Benyammi R, Moustafa K, Trémouillaux-Guiller J. Makhzoum A, et al. BioDrugs. 2014 Apr;28(2):145-59. doi: 10.1007/s40259-013-0062-1. BioDrugs. 2014. PMID: 23959796 Free PMC article. Review. - Downstream signaling mechanism of the C-terminal activation domain of transcriptional coactivator CoCoA.
Kim JH, Yang CK, Stallcup MR. Kim JH, et al. Nucleic Acids Res. 2006 May 22;34(9):2736-50. doi: 10.1093/nar/gkl361. Print 2006. Nucleic Acids Res. 2006. PMID: 16717280 Free PMC article. - Refining protein subcellular localization.
Scott MS, Calafell SJ, Thomas DY, Hallett MT. Scott MS, et al. PLoS Comput Biol. 2005 Nov;1(6):e66. doi: 10.1371/journal.pcbi.0010066. Epub 2005 Nov 25. PLoS Comput Biol. 2005. PMID: 16322766 Free PMC article.
References
- Adams, M.D., Dubnick, M., Kerlavage, A.R., Moreno, R., Kelley, J.M., Utterback, T.R., Nagle, J.W., Fields, C., and Venter, J.C. 1992. Sequence identification of 2,375 human brain genes. Nature 355: 632-634. - PubMed
- Aza-Blanc, P., Cooper, C.L., Wagner, K., Batalov, S., Deveraux, Q.L., and Cooke, M.P. 2003. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell 12: 627-637. - PubMed
WEB SITE REFERENCES
- http://genome.ucsc.edu/cgi-bin/hgGateway; UCSC Genome Browser GoldenPath.
- http://mips.gsf.de/projects/cdna; database with annotation of the cDNAs sequenced by the German cDNA Consortium.
- http://www.dkfz.de/LIFEdb; database with subcellular localizations and protein annotation (the address is case-sensitive).
- http://www.ebi.ac.uk/interpro/; IntroPro database of protein families, domains, and functional sites.
- http://www.ncbi.nlm.nih.gov/LocusLink/; LocusLink database with curated sequence and descriptive information on genetic loci.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials