Improvement of comparative model accuracy by free-energy optimization along principal components of natural structural variation - PubMed (original) (raw)
Comparative Study
. 2004 Oct 26;101(43):15346-51.
doi: 10.1073/pnas.0404703101. Epub 2004 Oct 18.
Affiliations
- PMID: 15492216
- PMCID: PMC524448
- DOI: 10.1073/pnas.0404703101
Comparative Study
Improvement of comparative model accuracy by free-energy optimization along principal components of natural structural variation
Bin Qian et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Accurate high-resolution refinement of protein structure models is a formidable challenge because of the delicate balance of forces in the native state, the difficulty in sampling the very large number of alternative tightly packed conformations, and the inaccuracies in current force fields. Indeed, energy-based refinement of comparative models generally leads to degradation rather than improvement in model quality, and, hence, most current comparative modeling procedures omit physically based refinement. However, despite their inaccuracies, current force fields do contain information that is orthogonal to the evolutionary information on which comparative models are based, and, hence, refinement might be able to improve comparative models if the space that is sampled is restricted sufficiently so that false attractors are avoided. Here, we use the principal components of the variation of backbone structures within a homologous family to define a small number of evolutionarily favored sampling directions and show that model quality can be improved by energy-based optimization along these directions.
Figures
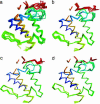
Fig. 1.
Variation in structure family and variation represented by PCs. (a) Superposition of the cores of all eight structures in structure family a.133.1.1. (b–d) Backbone conformations sampled along the direction defined by the first PC (b), the second PC (c), and the third PC (d).
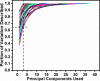
Fig. 2.
Fraction of structural variation explained by PCs. Each line represents a structure family. With additional PCs, more and more variation can be described. The dashed line indicates that the first three PCs can describe at least 65% of the variation observed in each structure family.
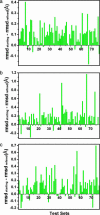
Fig. 3.
Improvement of protein backbone core region by sampling along the directions defined by the first three PCs and selecting low-energy decoys by using the Rosetta energy function. rmsd improvement of the lowest-energy decoys in tests I (a), II (b), and III (c) is shown. The improvement of rmsd is measured by (rmsd of starting model - rmsd of refined model). Positive values indicate improvement of backbone conformations.
Fig. 4.
Example of successful model refinement. Red, model structure; blue, native structure; green, refined structure. The rmsd between the model structure and native structure is 2.36 Å, and the rmsd between the refined structure and native structure is 1.42 Å.
Similar articles
- An efficient conformational sampling method for homology modeling.
Han R, Leo-Macias A, Zerbino D, Bastolla U, Contreras-Moreira B, Ortiz AR. Han R, et al. Proteins. 2008 Apr;71(1):175-88. doi: 10.1002/prot.21672. Proteins. 2008. PMID: 17985353 - Progress and challenges in high-resolution refinement of protein structure models.
Misura KM, Baker D. Misura KM, et al. Proteins. 2005 Apr 1;59(1):15-29. doi: 10.1002/prot.20376. Proteins. 2005. PMID: 15690346 - Refinement of NMR structures using implicit solvent and advanced sampling techniques.
Chen J, Im W, Brooks CL 3rd. Chen J, et al. J Am Chem Soc. 2004 Dec 15;126(49):16038-47. doi: 10.1021/ja047624f. J Am Chem Soc. 2004. PMID: 15584737 - A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction.
Moult J. Moult J. Curr Opin Struct Biol. 2005 Jun;15(3):285-9. doi: 10.1016/j.sbi.2005.05.011. Curr Opin Struct Biol. 2005. PMID: 15939584 Review. - Force fields for homology modeling.
Bordner AJ. Bordner AJ. Methods Mol Biol. 2012;857:83-106. doi: 10.1007/978-1-61779-588-6_4. Methods Mol Biol. 2012. PMID: 22323218 Review.
Cited by
- Cooperative dynamics of proteins unraveled by network models.
Eyal E, Dutta A, Bahar I. Eyal E, et al. Wiley Interdiscip Rev Comput Mol Sci. 2011 May-Jun;1(3):426-439. doi: 10.1002/wcms.44. Epub 2011 Apr 11. Wiley Interdiscip Rev Comput Mol Sci. 2011. PMID: 32148561 Free PMC article. - Homology modeling a fast tool for drug discovery: current perspectives.
Vyas VK, Ukawala RD, Ghate M, Chintha C. Vyas VK, et al. Indian J Pharm Sci. 2012 Jan;74(1):1-17. doi: 10.4103/0250-474X.102537. Indian J Pharm Sci. 2012. PMID: 23204616 Free PMC article. - vGNM: a better model for understanding the dynamics of proteins in crystals.
Song G, Jernigan RL. Song G, et al. J Mol Biol. 2007 Jun 8;369(3):880-93. doi: 10.1016/j.jmb.2007.03.059. Epub 2007 Mar 28. J Mol Biol. 2007. PMID: 17451743 Free PMC article. - Comparative modelling of protein structure and its impact on microbial cell factories.
Centeno NB, Planas-Iglesias J, Oliva B. Centeno NB, et al. Microb Cell Fact. 2005 Jun 30;4:20. doi: 10.1186/1475-2859-4-20. Microb Cell Fact. 2005. PMID: 15989691 Free PMC article. - Feature space resampling for protein conformational search.
Blum B, Jordan MI, Baker D. Blum B, et al. Proteins. 2010 May 1;78(6):1583-93. doi: 10.1002/prot.22677. Proteins. 2010. PMID: 20131376 Free PMC article.
References
- Sanchez, R., Pieper, U., Melo, F., Eswar, N., Marti-Renom, M. A., Madhusudhan, M. S., Mirkovic, N. & Sali, A. (2000) Nat. Struct. Biol. 7, Suppl., 986-990. - PubMed
- Baker, D. & Sali, A. (2001) Science 294, 93-96. - PubMed
- Stevens, R. C., Yokoyama, S. & Wilson, I. A. (2001) Science 294, 89-92. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources