Processing of bistranded abasic DNA clusters in gamma-irradiated human hematopoietic cells - PubMed (original) (raw)
. 2004 Oct 19;32(18):5609-20.
doi: 10.1093/nar/gkh871. Print 2004.
Collaborators, Affiliations
- PMID: 15494449
- PMCID: PMC524283
- DOI: 10.1093/nar/gkh871
Processing of bistranded abasic DNA clusters in gamma-irradiated human hematopoietic cells
Alexandros G Georgakilas et al. Nucleic Acids Res. 2004.
Abstract
Clustered DNA damages--two or more lesions on opposing strands and within one or two helical turns--are formed in cells by ionizing radiation or radiomimetic antitumor drugs. They are hypothesized to be difficult to repair, and thus are critical biological damages. Since individual abasic sites can be cytotoxic or mutagenic, abasic DNA clusters are likely to have significant cellular impact. Using a novel approach for distinguishing abasic clusters that are very closely spaced (putrescine cleavage) or less closely spaced (Nfo protein cleavage), we measured induction and processing of abasic clusters in 28SC human monocytes that were exposed to ionizing radiation. gamma-rays induced approximately 1 double-strand break: 1.3 putrescine-detected abasic clusters: 0.8 Nfo-detected abasic clusters. After irradiation, the 28SC cells rejoined double-strand breaks efficiently within 24 h. In contrast, in these cells, the levels of abasic clusters decreased very slowly over 14 days to background levels. In vitro repair experiments that used 28SC cell extracts further support the idea of slow processing of specific, closely spaced abasic clusters. Although some clusters were removed by active cellular repair, a substantial number was apparently decreased by 'splitting' during DNA replication and subsequent cell division. The existence of abasic clusters in 28SC monocytes, several days after irradiation suggests that they constitute persistent damages that could lead to mutation or cell killing.
Figures
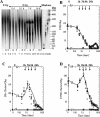
Figure 1
Abasic clusters in human monocytes exposed to 0–20 Gy of γ-rays. (A and B) Titration of human DNA cleavage by Nfo protein or PUTR. DNA in agarose plugs from cells irradiated with 0 or 20 Gy of γ-rays was treated with increasing amounts of Nfo protein (A) or PUTR (B) and the cluster frequencies determined. About 100 ng Nfo protein and 100 mM PUTR per 500 ng of DNA were chosen for all further experiments. The curves were fit to the points by eye. (C) DSB, (D) Nfo-abasic cluster and (E) PUTR-abasic cluster levels as a function of dose. Solid symbols correspond to individual data points and open symbols to averages. Error bars, SEM, in some cases are smaller than the corresponding symbol. Lines represent the least squares fit of the average points.
Figure 2
Processing of DNA lesions by human 28SC monocytes exposed to 5 Gy of γ-rays as a function of post-exposure time. (A) Electronic image of a neutral electrophoretic gel containing DNA from human 28SC monocytes exposed to 5 Gy of γ-rays, and harvested as a function of time post-exposure. 0 Gy, lanes 1–3; 5 Gy at increasing times (0–14 days), lanes 4–15; molecular size markers, lanes 16–19: T4, T7 and BglI digest of T7, showing here only the 22 kb fragment, lane 16; S.cerevisae, lane 17; H.wingei, lane 18; S.pombe, lane 19. The molecular sizes of these DNAs are shown in kb at the right of the gel image. For each time point three samples are shown: untreated, −; Nfo-treated, N; PUTR-treated, P. (B) DSB levels: 0 Gy, circles; 5 Gy, squares. (C) Nfo-detected abasic cluster levels: 0 Gy, circles; 5 Gy, diamonds; and (D) PUTR-detected cluster levels: 0 Gy, circles; 5 Gy, inverted triangles. Solid symbols correspond to individual data points and open symbols to average values. Error bars, SEM, where not shown are smaller than the corresponding symbol.
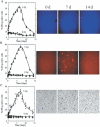
Figure 3
Staining of unirradiated or irradiated 28SC human monocytes by Hoechst 33258, propidium iodide, or erythrosin B. Percent of cells stained with (A) Hoechst 33258 (apoptotic cells, intense blue), (B) Propidium iodide (late apoptotic/dead cells, intense red), and (C) Erythrosin B (dead cells, intense black), after exposure of cells to 0 Gy (circles) or 5 Gy, (triangles) of γ-rays and incubation for the times indicated. Solid symbols correspond to individual data points and open symbols to average values. Error bars, SEM, where not shown are smaller than the corresponding symbol. The curves were fit to the points by eye. Images correspond to irradiated samples (5 Gy) at 0, 7, or 14 days after irradiation.
Figure 4
Growth of irradiated (5 Gy) and unirradiated 28SC cells, and potential effects on abasic cluster processing. (A) Cell concentration for 0–3 days after irradiation, during which the irradiated cells approximately doubled: 0 Gy (circles) 5 Gy, (triangles). Inset: cell concentration measured for 0–14 days. (B and C) Abasic cluster levels; experimental averages (open symbols), calculated for ‘splitting effect’ (solid symbols); (B) Nfo protein, diamonds and (C) putrescine, squares. Curves, power best fittings (y = axb) for the experimental points (solid lines), Nfo protein, a = 47.21 b = 0.60; PUTR, a = 67.04, b = 0.56; fit to the points by eye (dotted lines) for the calculated points. Solid symbols correspond to individual data points and open symbols to average values. Error bars, SEM, where not shown are smaller than the corresponding symbol.
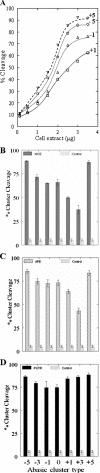
Figure 5
Cleavage of abasic clusters by extracts of 28SC cells, PUTR or hAPE1. (A) Oligonucleotides (23 bp) containing bistranded AP sites at different interlesion spacings (Table 1) were incubated (37°C, 12 min) with increasing quantities of extract. Cleavage efficiency is given as the percentage of conversion of the intact oligonucleotides to specific product fragments of lower size. (B) Cleavage efficiency by cell extract (WCE, 3 μg), (C) hAPE1 (2.5 ng) or (D) PUTR (1 mM) of specific abasic clusters (Table 1). The controls reflect the average ‘background’ cleavage by the extract of the control oligonucleotide containing only one abasic site (see Table 1). In neutral electrophoretic gels, no double-strand cleavage is expected for an oligonucleotide containing a single AP site. Similar cleavage was found for the substrate containing a single abasic site by PUTR and hAPE1 (data not shown).
Similar articles
- High efficiency detection of bi-stranded abasic clusters in gamma-irradiated DNA by putrescine.
Georgakilas AG, Bennett PV, Sutherland BM. Georgakilas AG, et al. Nucleic Acids Res. 2002 Jul 1;30(13):2800-8. doi: 10.1093/nar/gkf393. Nucleic Acids Res. 2002. PMID: 12087163 Free PMC article. - Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation.
Sutherland BM, Bennett PV, Sidorkina O, Laval J. Sutherland BM, et al. Proc Natl Acad Sci U S A. 2000 Jan 4;97(1):103-8. doi: 10.1073/pnas.97.1.103. Proc Natl Acad Sci U S A. 2000. PMID: 10618378 Free PMC article. - Low levels of endogenous oxidative damage cluster levels in unirradiated viral and human DNAs.
Sutherland BM, Bennett PV, Cintron NS, Guida P, Laval J. Sutherland BM, et al. Free Radic Biol Med. 2003 Sep 1;35(5):495-503. doi: 10.1016/s0891-5849(03)00327-7. Free Radic Biol Med. 2003. PMID: 12927599 - Securing genome stability by orchestrating DNA repair: removal of radiation-induced clustered lesions in DNA.
Dianov GL, O'Neill P, Goodhead DT. Dianov GL, et al. Bioessays. 2001 Aug;23(8):745-9. doi: 10.1002/bies.1104. Bioessays. 2001. PMID: 11494323 Review. - Clustered DNA damages as dosemeters for ionising radiation exposure and biological responses.
Sutherland BM, Bennett PV, Saparbaev M, Sutherland JC, Laval J. Sutherland BM, et al. Radiat Prot Dosimetry. 2001;97(1):33-8. doi: 10.1093/oxfordjournals.rpd.a006635. Radiat Prot Dosimetry. 2001. PMID: 11763355 Review.
Cited by
- Histone-catalyzed cleavage of nucleosomal DNA containing 2-deoxyribonolactone.
Zhou C, Greenberg MM. Zhou C, et al. J Am Chem Soc. 2012 May 16;134(19):8090-3. doi: 10.1021/ja302993h. Epub 2012 May 2. J Am Chem Soc. 2012. PMID: 22551239 Free PMC article. - DNA Damage Response and Repair in Boron Neutron Capture Therapy.
Mechetin GV, Zharkov DO. Mechetin GV, et al. Genes (Basel). 2023 Jan 2;14(1):127. doi: 10.3390/genes14010127. Genes (Basel). 2023. PMID: 36672868 Free PMC article. Review. - 5-Iodo-4-thio-2'-Deoxyuridine as a Sensitizer of X-ray Induced Cancer Cell Killing.
Makurat S, Spisz P, Kozak W, Rak J, Zdrowowicz M. Makurat S, et al. Int J Mol Sci. 2019 Mar 15;20(6):1308. doi: 10.3390/ijms20061308. Int J Mol Sci. 2019. PMID: 30875879 Free PMC article. - Molecular analysis of base damage clustering associated with a site-specific radiation-induced DNA double-strand break.
Datta K, Jaruga P, Dizdaroglu M, Neumann RD, Winters TA. Datta K, et al. Radiat Res. 2006 Nov;166(5):767-81. doi: 10.1667/RR0628.1. Radiat Res. 2006. PMID: 17067210 Free PMC article.
References
- Ward J.F. (1981) Some biochemical consequences of the spatial distribution of ionizing radiation produced free radicals. Radiat. Res., 86, 185–195. - PubMed
- Goodhead D.T. (1994) Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int. J. Rad. Biol., 65, 7–17. - PubMed
- Ahnstrom G. and Bryant,P.E. (1982) DNA double-strand breaks generated by the repair of X-ray damage in Chinese hamster cells. Int. J. Radiat. Biol., 41, 671–676. - PubMed