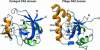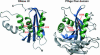RNAi: finding the elusive endonuclease - PubMed (original) (raw)
Review
RNAi: finding the elusive endonuclease
Andreas Lingel et al. RNA. 2004 Nov.
Abstract
RNA interference involves endonucleolytic cleavage of mRNAs at a site determined by complementary siRNAs. Initial cleavage leads to rapid degradation of the message, resulting in a corresponding reduction in the level of the encoded protein. Despite intensive study, the identity of the endonucleolytic activity (designated slicer) has remained obscure. Now, a combination of structural and biochemical analyses provide compelling evidence that human Argonaute2 (Ago2), a protein already known to be a key player in the RNAi pathway, is in fact the missing endonuclease.
Figures
FIGURE 1.
Comparison of the Drosophila melanogaster Argonaute2 PAZ domain (DmAgo2, left) and the PAZ domain of the Argonaute protein from Pyrococcus furiosus (PfAgo, right). Structures are shown as ribbon diagrams, with helices colored green and β-strands shown in blue. The β-hairpin/α-helix module is shown in orange. This module contains two β-helices in the PAZ domain of PfAgo. Side chains of residues shown to contact nucleic acids in structures of the DmAgo2 PAZ domain in complex with nucleic acids are shown in gray and are labeled (Lingel et al. 2004). Residues in equivalent positions in the PfAgo protein are indicated.
FIGURE 2.
The Piwi domain of the PfAgo protein comprises an RNase H fold. The structure of RNase HI from E. coli is shown on the left (RCSB protein data bank accession no. 1RDD) (Katayanagi et al. 1993), the Piwi domain of the PfAgo protein on the right. Secondary structure elements that constitute the RNase H fold are colored as in Figure 1 ▶; additional structural elements are shown in gray. Catalytic side chains of RNase HI and the corresponding side chains in the PfAgo protein are indicated and labeled. Figures 1 ▶ and 2 ▶ were prepared with the programm MOLMOL (Koradi et al. 1996).
Similar articles
- The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes.
Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. Song JJ, et al. Nat Struct Biol. 2003 Dec;10(12):1026-32. doi: 10.1038/nsb1016. Epub 2003 Nov 16. Nat Struct Biol. 2003. PMID: 14625589 - Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain.
Lingel A, Simon B, Izaurralde E, Sattler M. Lingel A, et al. Nature. 2003 Nov 27;426(6965):465-9. doi: 10.1038/nature02123. Epub 2003 Nov 16. Nature. 2003. PMID: 14615801 - Dicing and slicing: the core machinery of the RNA interference pathway.
Hammond SM. Hammond SM. FEBS Lett. 2005 Oct 31;579(26):5822-9. doi: 10.1016/j.febslet.2005.08.079. Epub 2005 Sep 27. FEBS Lett. 2005. PMID: 16214139 Review. - Short interfering RNA strand selection is independent of dsRNA processing polarity during RNAi in Drosophila.
Preall JB, He Z, Gorra JM, Sontheimer EJ. Preall JB, et al. Curr Biol. 2006 Mar 7;16(5):530-5. doi: 10.1016/j.cub.2006.01.061. Curr Biol. 2006. PMID: 16527750 - The Argonautes.
Joshua-Tor L. Joshua-Tor L. Cold Spring Harb Symp Quant Biol. 2006;71:67-72. doi: 10.1101/sqb.2006.71.048. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381282 Review.
Cited by
- Genome-Wide Identification and Characterization of Major RNAi Genes Highlighting Their Associated Factors in Cowpea (Vigna unguiculata (L.) Walp.).
Hasan MN, Mosharaf MP, Uddin KS, Das KR, Sultana N, Noorunnahar M, Naim D, Mollah MNH. Hasan MN, et al. Biomed Res Int. 2023 Nov 24;2023:8832406. doi: 10.1155/2023/8832406. eCollection 2023. Biomed Res Int. 2023. PMID: 38046903 Free PMC article. - Alternative initiation and splicing in dicer gene expression in human breast cells.
Irvin-Wilson CV, Chaudhuri G. Irvin-Wilson CV, et al. Breast Cancer Res. 2005;7(4):R563-9. doi: 10.1186/bcr1043. Epub 2005 May 16. Breast Cancer Res. 2005. PMID: 15987463 Free PMC article. - Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence.
Koonin EV. Koonin EV. Biol Direct. 2017 Feb 10;12(1):5. doi: 10.1186/s13062-017-0177-2. Biol Direct. 2017. PMID: 28187792 Free PMC article. Review. - In silico identification and characterization of AGO, DCL and RDR gene families and their associated regulatory elements in sweet orange (Citrus sinensis L.).
Mosharaf MP, Rahman H, Ahsan MA, Akond Z, Ahmed FF, Islam MM, Moni MA, Mollah MNH. Mosharaf MP, et al. PLoS One. 2020 Dec 21;15(12):e0228233. doi: 10.1371/journal.pone.0228233. eCollection 2020. PLoS One. 2020. PMID: 33347517 Free PMC article.
References
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. - PubMed
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. - PubMed
- Carmell, M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. 2002. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 16: 2733–2742. - PubMed
- Caudy, A.A., Ketting, R.F., Hammond, S.M., Denli, A.M., Bathoorn, A.M., Tops, B.B., Silva, J.M., Myers, M.M., Hannon, G.J., and Plasterk, R.H. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414. - PubMed
- Cerutti, L., Mian, N., and Bateman, A. 2000. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25: 481–482. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources