Silencing of SARS-CoV spike gene by small interfering RNA in HEK 293T cells - PubMed (original) (raw)
Silencing of SARS-CoV spike gene by small interfering RNA in HEK 293T cells
Zhao-ling Qin et al. Biochem Biophys Res Commun. 2004.
Abstract
Two candidate small interfering RNAs (siRNAs) corresponding to severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike gene were designed and in vitro transcribed to explore the possibility of silencing SARS-CoV S gene. The plasmid pEGFP-optS, which contains the codon-optimized SARS-CoV S gene and expresses spike-EGFP fusion protein (S-EGFP) as silencing target and expressing reporter, was transfected with siRNAs into HEK 293T cells. At various time points of posttransfection, the levels of S-EGFP expression and amounts of spike mRNA transcript were detected by fluorescence microscopy, flow cytometry, Western blot, and real-time quantitative PCR, respectively. The results showed that the cells transfected with pEGFP-optS expressed S-EGFP fusion protein at a higher level compared with those transfected with pEGFP-S, which contains wildtype SARS-CoV spike gene sequence. The green fluorescence, mean fluorescence intensity, and SARS-CoV S RNA transcripts were found significantly reduced, and the expression of SARS-CoV S glycoprotein was strongly inhibited in those cells co-transfected with either EGFP- or S-specific siRNAs. Our findings demonstrated that the S-specific siRNAs used in this study were able to specifically and effectively inhibit SARS-CoV S glycoprotein expression in cultured cells through blocking the accumulation of S mRNA, which may provide an approach for studies on the functions of SARS-CoV S gene and development of novel prophylactic or therapeutic agents for SARS-CoV.
Figures
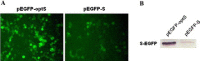
Fig. 1
Differences in the efficiency of expression of modified and unmodified SARS-CoV spike genes. (A) Enhanced fluorescence imaging in codon-optimized SARS-CoV spike gene transfected HEK 293T cells (left panel). (B) Western blot analysis of S-EGFP fusion protein expression in the HEK 293T cells transfected with pEGFP-optS or pEGFP-S.
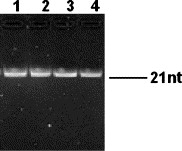
Fig. 2
Purity and integrity of the siRNAs transcribed in vitro. 1. EGFP-specific siRNA. 2. Scramble siRNA. 3. SARS-CoV S-siRNA1. 4. SARS-CoV S-siRNA2.
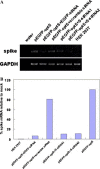
Fig. 3
Reduced SARS-CoV spike transcripts in S-specific siRNA transfected HEK 293T cells. (A) RT-PCR was performed to show the degradation of S mRNA in S-specific siRNA transfected cells. RT-PCR products for the spike gene and the internal control, GAPDH, are shown in the figure. (B) Real-time quantitative PCR was performed for spike and GAPDH RNA content on each sample. The assay shows the percentage of SARS-CoV spike mRNA in siRNA transfected HEK 293T cells relative to that in mock-transfected cells after being normalized to GAPDH mRNA.
Fig. 4
Effect of siRNAs on the expression of S-EGFP in HEK 293T cells. (A) HEK 293T cells. (B) pEGFP-optS transfected. (C) pEGFP-optS and EGFP siRNA co-transfected. (D) pEGFP-optS and scramble siRNA co-transfected. (E) pEGFP-optS and S-siRNA1 co-transfected. (F) pEGFP-optS and S-siRNA2 co-transfected. Upper panels represent the cell fluorescence images recorded at 48 h posttransfection. Lower panels represent the light microscopic view of cells in the same field. Specific silencing of the S-EGFP fusion protein expression was confirmed in three independent experiments.
Fig. 5
Flow cytometry analysis of EGFP expression in HEK 293T cells. (A–F) show the results from HEK 293T cells control, cells transfected with pEGFP-optS, cells co-transfected with pEGFP-optS and EGFP siRNA, cells co-transfected with pEGFP-optS and scramble siRNA, cells co-transfected with pEGFP-optS and S-siRNA1 and cells co-transfected with pEGFP-optS and S-siRNA2, respectively. At 48 h after transfection, cells were analyzed for EGFP expression by flow cytometry. The percentage of the cell population that exceeded the fluorescence intensity of the control cells and the mean fluorescence intensity of this population were calculated.
Fig. 6
Effect of siRNA on S-EGFP protein expression in HEK 293T cells. Western blot analysis was performed on equal amounts of proteins harvested from mock- or siRNA co-transfected HEK 293T cells at 48 h posttransfection by using GFP- and GAPDH-specific antibodies described in Materials and methods. β-Actin was used as a loading control.
Similar articles
- siRNAs targeting terminal sequences of the SARS-associated coronavirus membrane gene inhibit M protein expression through degradation of M mRNA.
Qin ZL, Zhao P, Cao MM, Qi ZT. Qin ZL, et al. J Virol Methods. 2007 Nov;145(2):146-54. doi: 10.1016/j.jviromet.2007.05.017. Epub 2007 Jun 27. J Virol Methods. 2007. PMID: 17590445 Free PMC article. - RNA interference effectively inhibits mRNA accumulation and protein expression of hepatitis C virus core and E2 genes in human cells.
Liu M, Ding H, Zhao P, Qin ZL, Gao J, Cao MM, Luan J, Wu WB, Qi ZT. Liu M, et al. Biosci Biotechnol Biochem. 2006 Sep;70(9):2049-55. doi: 10.1271/bbb.60001. Epub 2006 Sep 7. Biosci Biotechnol Biochem. 2006. PMID: 16960393 - Inhibition of SARS-CoV replication by siRNA.
Wu CJ, Huang HW, Liu CY, Hong CF, Chan YL. Wu CJ, et al. Antiviral Res. 2005 Jan;65(1):45-8. doi: 10.1016/j.antiviral.2004.09.005. Antiviral Res. 2005. PMID: 15652970 Free PMC article. - The spike protein of SARS-CoV--a target for vaccine and therapeutic development.
Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. Du L, et al. Nat Rev Microbiol. 2009 Mar;7(3):226-36. doi: 10.1038/nrmicro2090. Epub 2009 Feb 9. Nat Rev Microbiol. 2009. PMID: 19198616 Free PMC article. Review. - Expression, glycosylation, and modification of the spike (S) glycoprotein of SARS CoV.
Shen S, Tan TH, Tan YJ. Shen S, et al. Methods Mol Biol. 2007;379:127-35. doi: 10.1007/978-1-59745-393-6_9. Methods Mol Biol. 2007. PMID: 17502675 Free PMC article. Review.
Cited by
- Application of Nanotechnology in the COVID-19 Pandemic.
Yang D. Yang D. Int J Nanomedicine. 2021 Jan 26;16:623-649. doi: 10.2147/IJN.S296383. eCollection 2021. Int J Nanomedicine. 2021. PMID: 33531805 Free PMC article. Review. - Prospects for RNAi Therapy of COVID-19.
Uludağ H, Parent K, Aliabadi HM, Haddadi A. Uludağ H, et al. Front Bioeng Biotechnol. 2020 Jul 30;8:916. doi: 10.3389/fbioe.2020.00916. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32850752 Free PMC article. Review. - Antisense technology as a potential strategy for the treatment of coronaviruses infection: With focus on COVID-19.
Alavizadeh SH, Doagooyan M, Zahedipour F, Torghabe SY, Baharieh B, Soleymani F, Gheybi F. Alavizadeh SH, et al. IET Nanobiotechnol. 2022 May;16(3):67-77. doi: 10.1049/nbt2.12079. Epub 2022 Mar 10. IET Nanobiotechnol. 2022. PMID: 35274474 Free PMC article. Review. - PTEN Lipid Phosphatase Activity Enhances Dengue Virus Production through Akt/FoxO1/Maf1 Signaling.
Liu B, Gao TT, Fu XY, Xu ZH, Ren H, Zhao P, Qi ZT, Qin ZL. Liu B, et al. Virol Sin. 2021 Jun;36(3):412-423. doi: 10.1007/s12250-020-00291-6. Epub 2020 Oct 12. Virol Sin. 2021. PMID: 33044659 Free PMC article. - Emerging Technologies for the Treatment of COVID-19.
Aghamollaei H, Sarvestani R, Bakherad H, Zare H, Guest PC, Ranjbar R, Sahebkar A. Aghamollaei H, et al. Adv Exp Med Biol. 2021;1321:81-96. doi: 10.1007/978-3-030-59261-5_7. Adv Exp Med Biol. 2021. PMID: 33656715
References
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. SARS study group. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. - PMC - PubMed
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous