TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences - PubMed (original) (raw)
Comparative Study
TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences
Hanno Teeling et al. BMC Bioinformatics. 2004.
Abstract
Background: In the emerging field of environmental genomics, direct cloning and sequencing of genomic fragments from complex microbial communities has proven to be a valuable source of new enzymes, expanding the knowledge of basic biological processes. The central problem of this so called metagenome-approach is that the cloned fragments often lack suitable phylogenetic marker genes, rendering the identification of clones that are likely to originate from the same genome difficult or impossible. In such cases, the analysis of intrinsic DNA-signatures like tetranucleotide frequencies can provide valuable hints on fragment affiliation. With this application in mind, the TETRA web-service and the TETRA stand-alone program have been developed, both of which automate the task of comparative tetranucleotide frequency analysis.
Availability: http://www.megx.net/tetra.
Results: TETRA provides a statistical analysis of tetranucleotide usage patterns in genomic fragments, either via a web-service or a stand-alone program. With respect to discriminatory power, such an analysis outperforms the assignment of genomic fragments based on the (G+C)-content, which is a widely-used sequence-based measure for assessing fragment relatedness. While the web-service is restricted to the calculation of correlation coefficients between tetranucleotide usage patterns of submitted DNA sequences, the stand-alone program generates a much more detailed output, comprising all raw data and graphical plots. The stand-alone program is controlled via a graphical user interface and can batch-process a multitude of sequences. Furthermore, it comes with pre-computed tetranucleotide usage patterns for 166 prokaryote chromosomes, providing a useful reference dataset and source for data-mining.
Conclusions: Up to now, the analysis of skewed oligonucleotide distributions within DNA sequences is not a commonly used tool within metagenomics. With the TETRA web-service and stand-alone program, the method is now accessible in an easy to use manner for a broad audience. This will hopefully facilitate the interrelation of genomic fragments from metagenome libraries, ultimately leading to new insights into the genetic potentials of yet uncultured microorganisms.
Figures
Figure 1
Data-entry page of the TETRA web-service.
Figure 2
(left) Navigator window of the TETRA stand-alone program showing a subset of the pre-computed 166 prokaryote chromosomes for selection. (right) Data window with a subset of the tetranucleotide frequencies of the Aeropyrum pernix K1 chromosome.
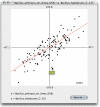
Figure 3
Dotplot of Z-scores for the tetranucleotide frequencies of two Bacilli. A regression line is calculated as well as the Pearson correlation coefficient (0.823). Hovering with the mouse over a dot shows a small information window with the tetranucleotide(s) the spot is referring to (as shown for GATC in this case).
References
- Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/AEM.66.6.2541-2547.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources